Primers, method and kit for whole genome detection of human mitochondria
A whole-genome and mitochondrial technology, applied in the field of gene mutation detection in biotechnology, can solve the problems of low sensitivity, long cycle, high cost, etc., and achieve the effects of improving primer specificity, simple operation, and cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A series of original templates with different mutation frequencies were used to simulate a series of original templates with known site mutations to verify the performance of this kit.
[0030] A. Template preparation for different mutation ratios
[0031] Plasmid DNA mutated at known sites (chrMT:11778G>A, GRCh37.p13) was mixed with wild-type DNA in proportion to obtain original templates with mutation frequencies of 0.05%, 1%, 2%, 5%, and 10%, respectively .
[0032] B. Multiplex PCR method to amplify the target region of the sample genome
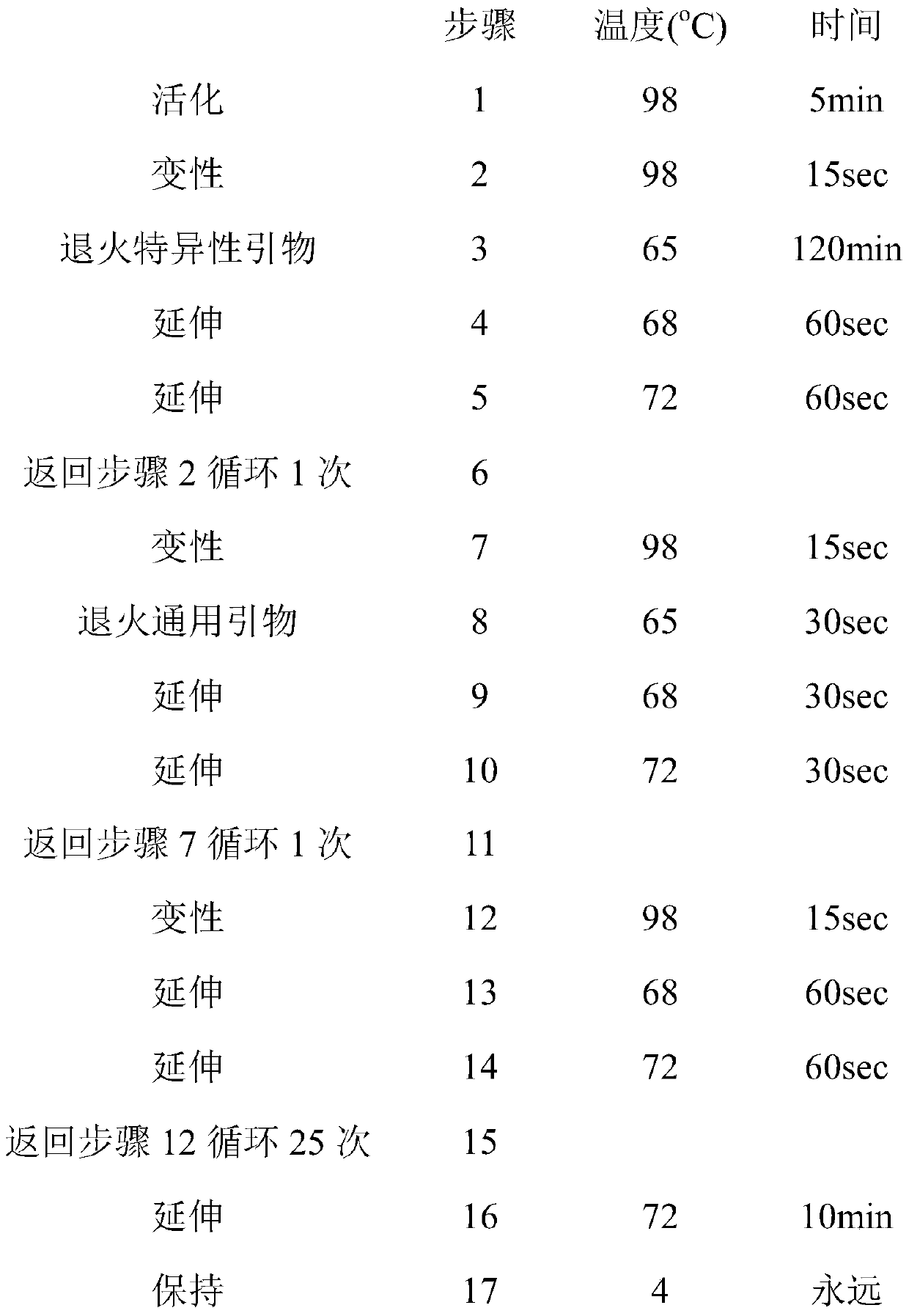
[0033] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare a PCR reaction system as shown in the table below. The amount of template for each reaction is 10ng, and be careful to avoid cross-infection.
[0034] components volume 2×PCR mix 6.25 μL specific primer 0.5μL Universal Primer R5 0.5μL Universal Primer R7 0.5μL DNA template, 50ng 1μL nuclease...
Embodiment 2
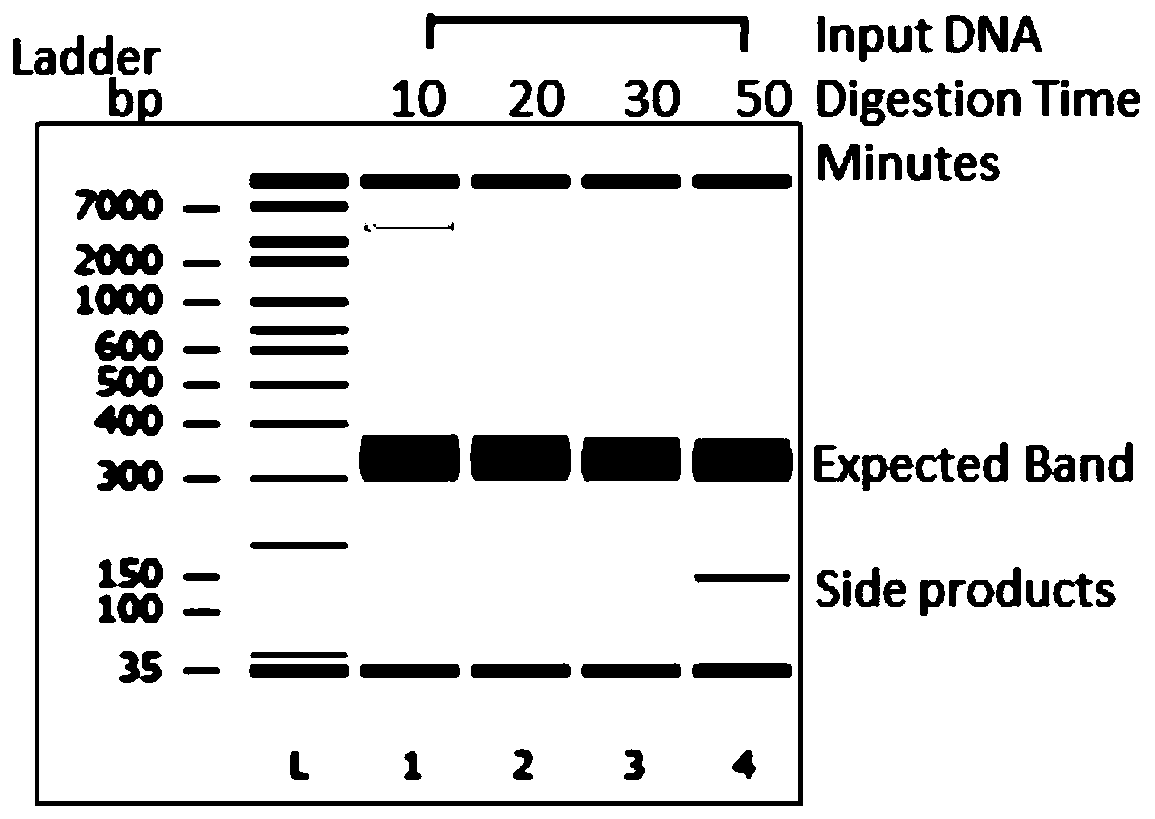
[0065] Genomic DNA with different degrees of degradation was used as a template to analyze the correlation between the sequencing results to verify the performance of the kit.
[0066] A. Genomic DNA preparation with different degrees of degradation
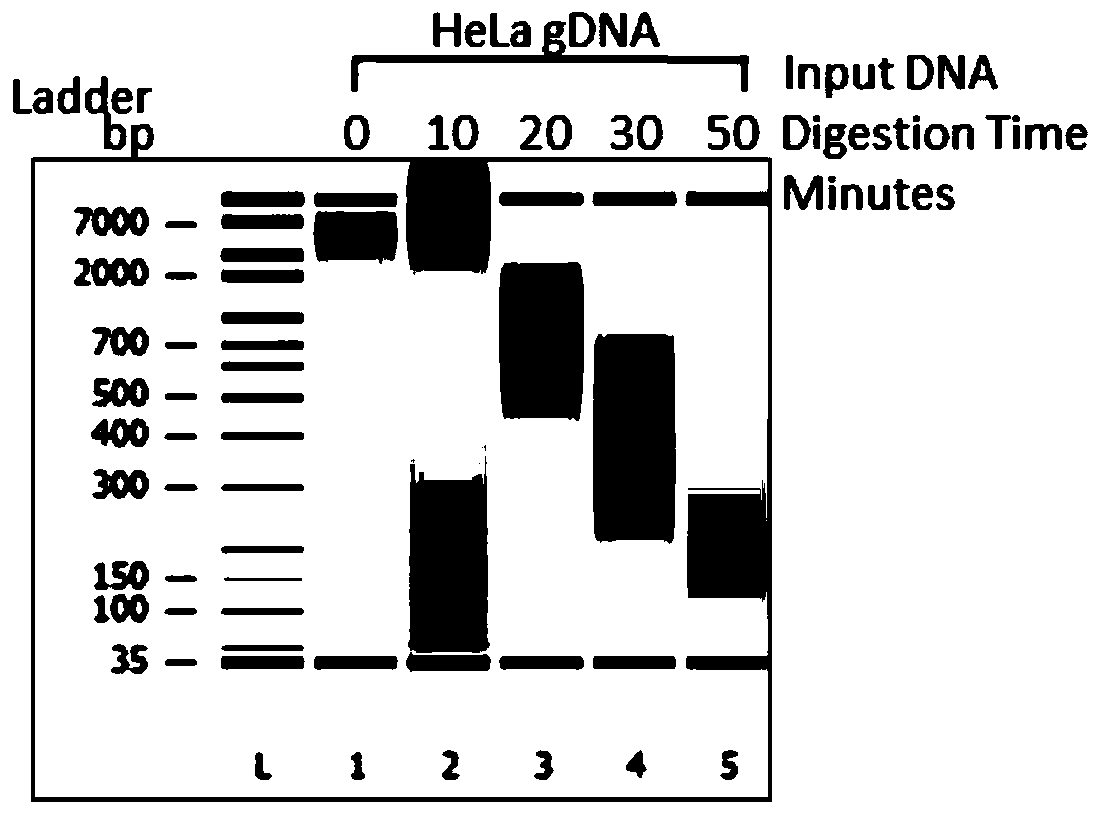
[0067] Using 250ng Hela genomic DNA (NEB, N4006S) as raw material, use dsDNAFragmentase (NEB, M0348S) digested the raw materials at 37°C for 0, 10, 20, 30, 50min and other different times, and the digested products were processed by Agilent 2100 instruments for quantitative and quality inspection. Such as figure 1 As shown, the raw material was obviously degraded after being digested at 37°C for 20 minutes, and the longer the treatment time, the more serious the degradation.
[0068] B. Multiplex PCR method to amplify the target region of the sample genome
[0069] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare a PCR reaction system as shown in the table below. The amount of template for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com