Flight mass spectrometry genotyping detection method
A genotyping and flight mass spectrometry technology, which is applied in the field of flight mass spectrometry genotyping and detection, can solve problems such as weak signals of high molecular weight products, and achieve the effects of improving detection resolution, high throughput and high resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1: Designing 4 probes related to the risk of type 2 diabetes
[0043] Type 2 diabetes (type2diabetes, T2D) is a polygenic genetic disease. Its clinical diagnosis is mainly evaluated by the corresponding indicators of glycosylated hemoglobin, fasting blood glucose and glucose tolerance. It cannot be detected and diagnosed as early as possible, thereby affecting early intervention and treatment by mistake, and bringing greater pain and economic loss to patients. If the etiology of the disease is combined with personalized molecular testing, early diagnosis of T2D can be realized. Combined with domestic and foreign genome-wide association and candidate gene methods, the T2D single nucleotide polymorphic sites reported are relatively high in the Chinese population. 4 SNPs highly correlated with T2D susceptibility were selected.
[0044] For the selected 4 SNPs, the upstream and downstream flanking sequences of the SNP sites were obtained from the 1000Genomes websit...
Embodiment 2
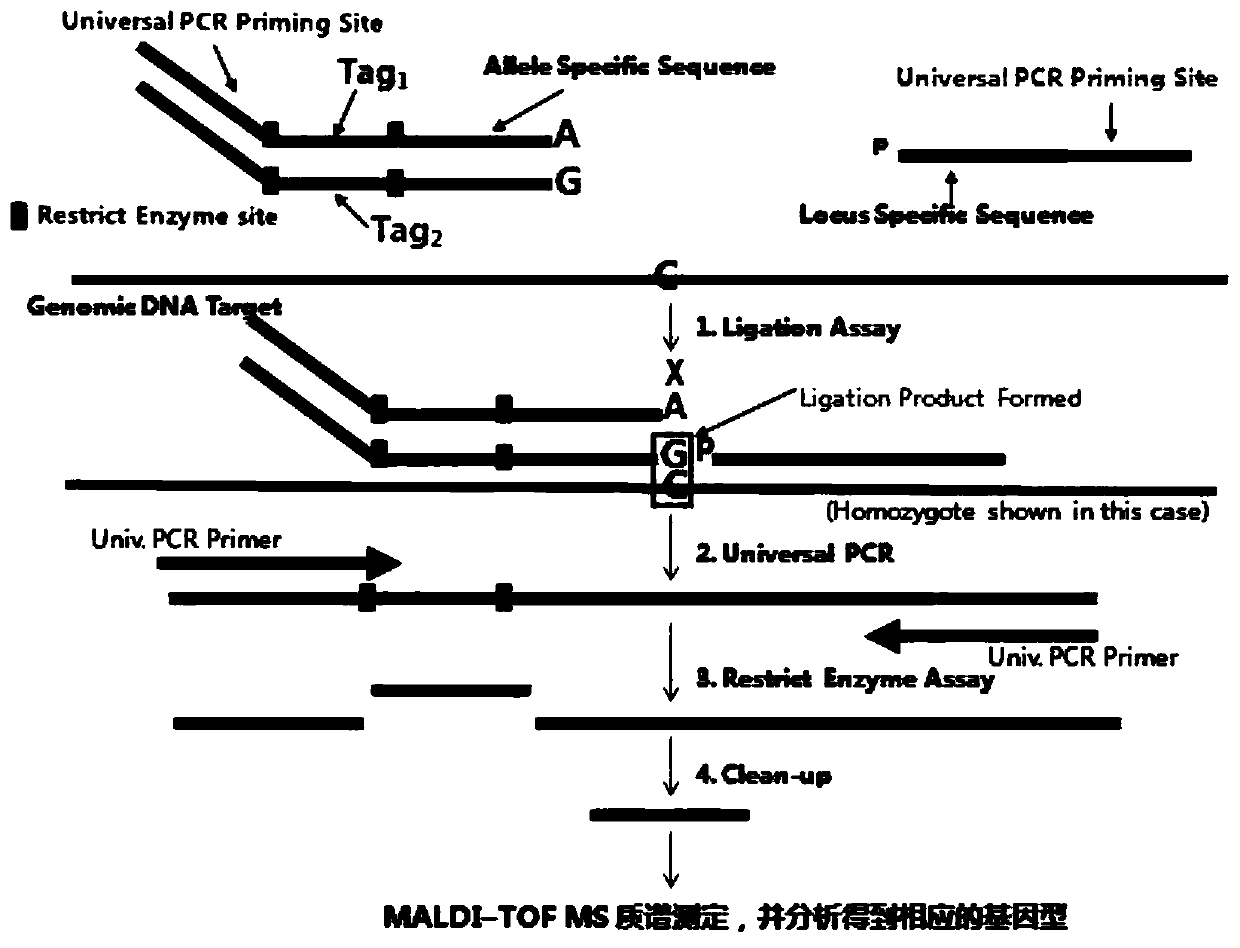
[0046] Example 2: Oligonucleotide Ligation Reaction (OLA) and Enrichment of Ligation Products
[0047] Fragment the sample genomic DNA, adjust the sample genomic DNA concentration to 100ng / μl with 1×TE (10mM Tris-HCL PH8.0,0.1mM EDTA), take out 15μl, 99℃ for 6 minutes, so that the DNA fragments are distributed in 2-7kb between.
[0048] The reaction system for configuring OLA is 0.3μl OLA buffer (10×), 0.3μl DTT (25mM), 0.04μl TaqDNA Ligase (40U / μl), 0.03μl ASO Oligo Mix, 0.03μl LSO Oligo Mix, 2.3μl ddH2O. And take 3 μ l of the above reaction system and divide it into the centrifuge tube that has added the fragmented sample, and carry out the oligonucleotide ligation reaction. The reaction conditions were: denaturation at 95°C for 5 minutes, 3 cycles (95°C for 30s and 50°C for 25 minutes), and storage at 4°C.
[0049] Use general-purpose PCR primers Forword-(5 / -CCCAGTCACGACGTTGTAAAACG-3 / ), Reverse (5 / -AGCGGATAACAATTTCACACAGG-3 / ) to amplify the above ligation products. The PC...
Embodiment 3
[0050] Example 3: Restriction enzyme reaction and purification of digested products
[0051] According to different restriction endonucleases, the reaction system and reaction conditions will be different. In this example, DpnI restriction endonuclease and its corresponding buffer were added to the above PCR product for digestion. The digestion system was: 15 μl of PCR product, 2 μl of Buffer (10×), 1 μl of DpnI, and 4 μl of ddH2O. After digestion at 37°C for 4 hours, the PCR product will be cut into multiple fragments, and the fragments carrying Tags are biotin-labeled, so they can be purified using streptavidin-coated beads to retain biotin-coated beads. Modified Tags sequence. Take an equal volume of the digested product and add it to the resuspended streptavidin magnetic beads, shake well, shake gently at room temperature for 30 minutes, put the centrifuge tube on the magnetic stand, absorb for about 30 seconds, and discard the supernatant. Remove the centrifuge tube fro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com