Saccharomyces cerevisiae engineering bacteria for producing dihydroarteannuic acid and construction method and application thereof
A technology of dihydroartemisinic acid and Saccharomyces cerevisiae, which is applied in the field of microorganisms, can solve the problems of high proportion of artemisinic acid, influence the generation of target substances, etc., and achieve the effect of increasing the proportion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
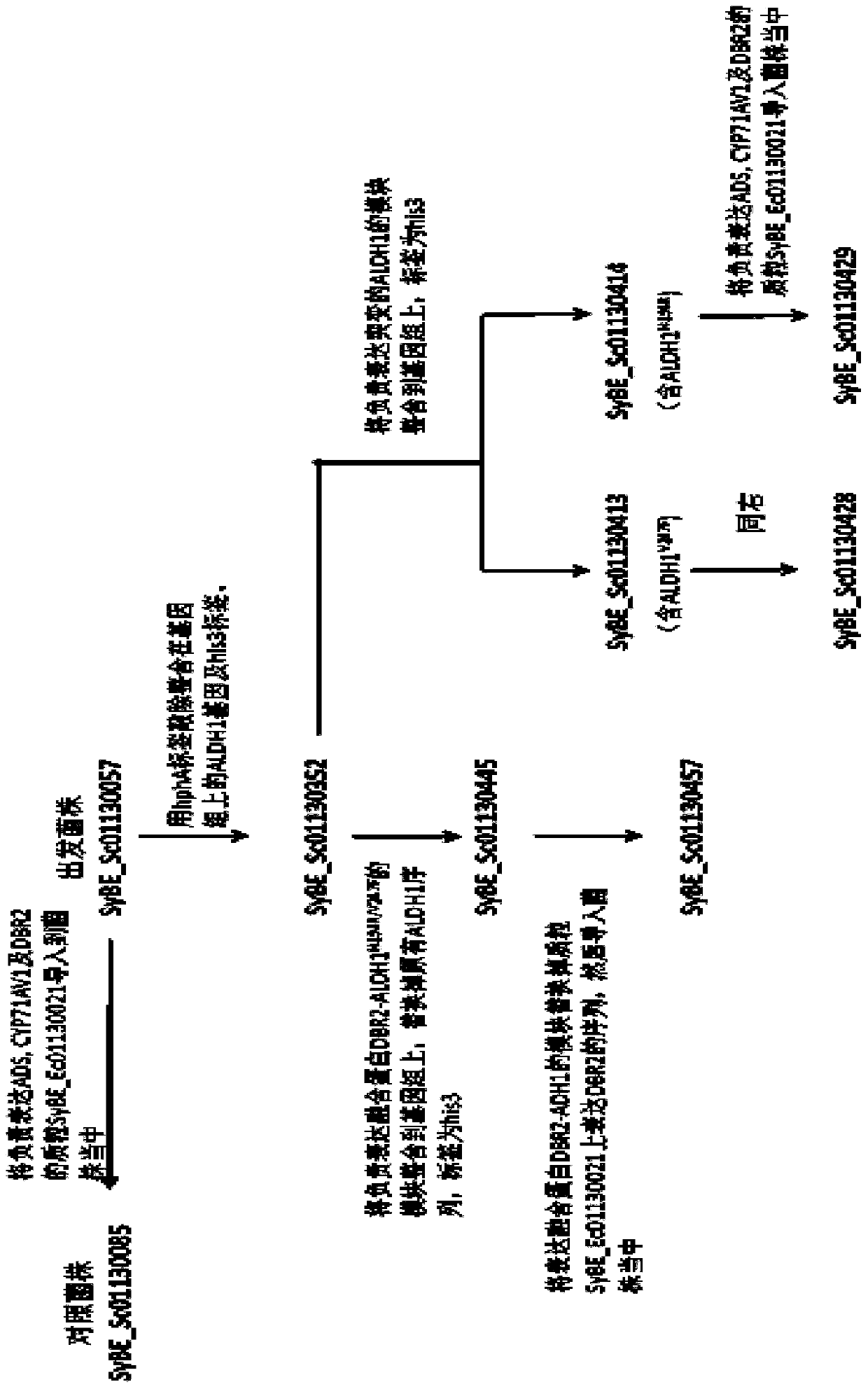
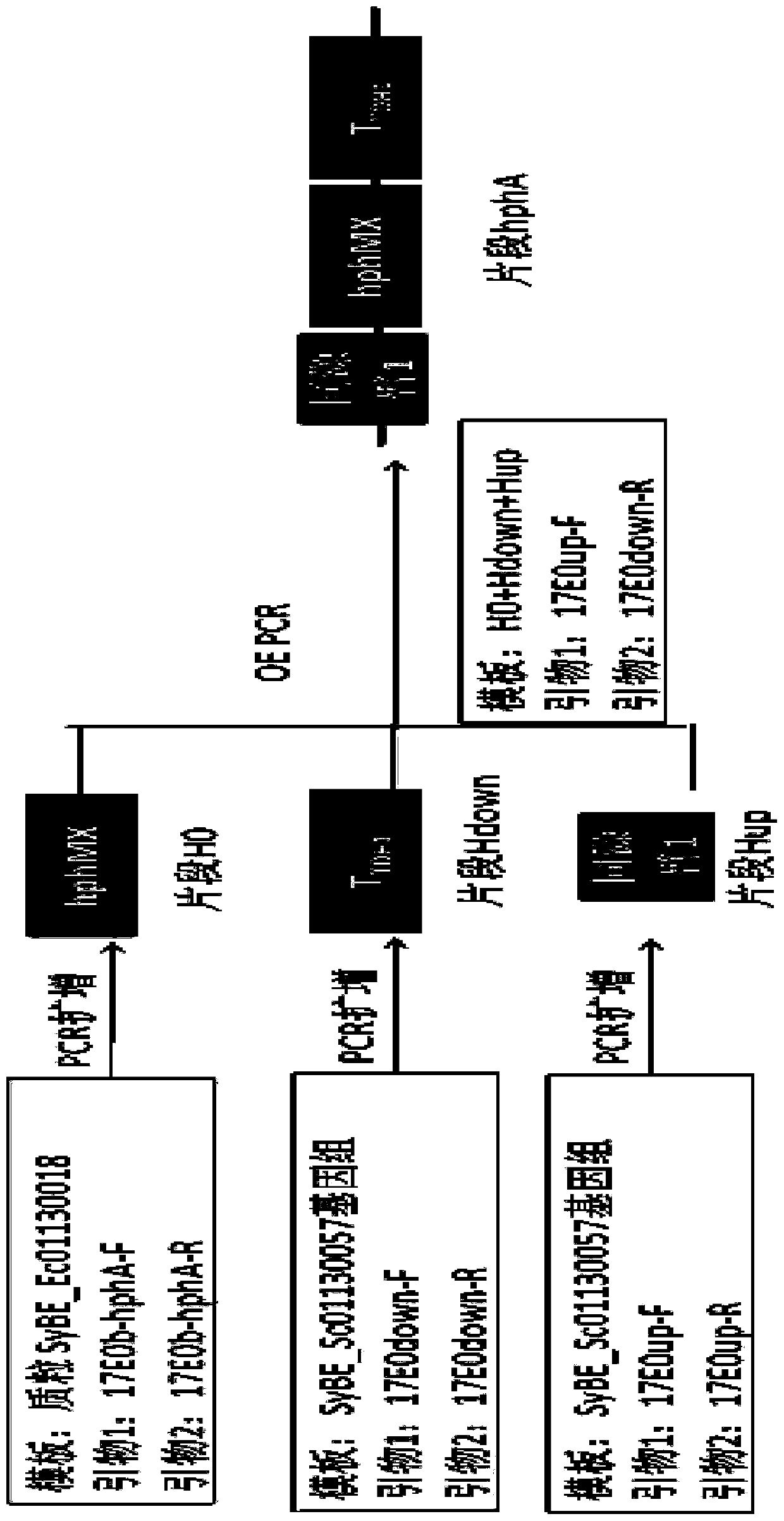
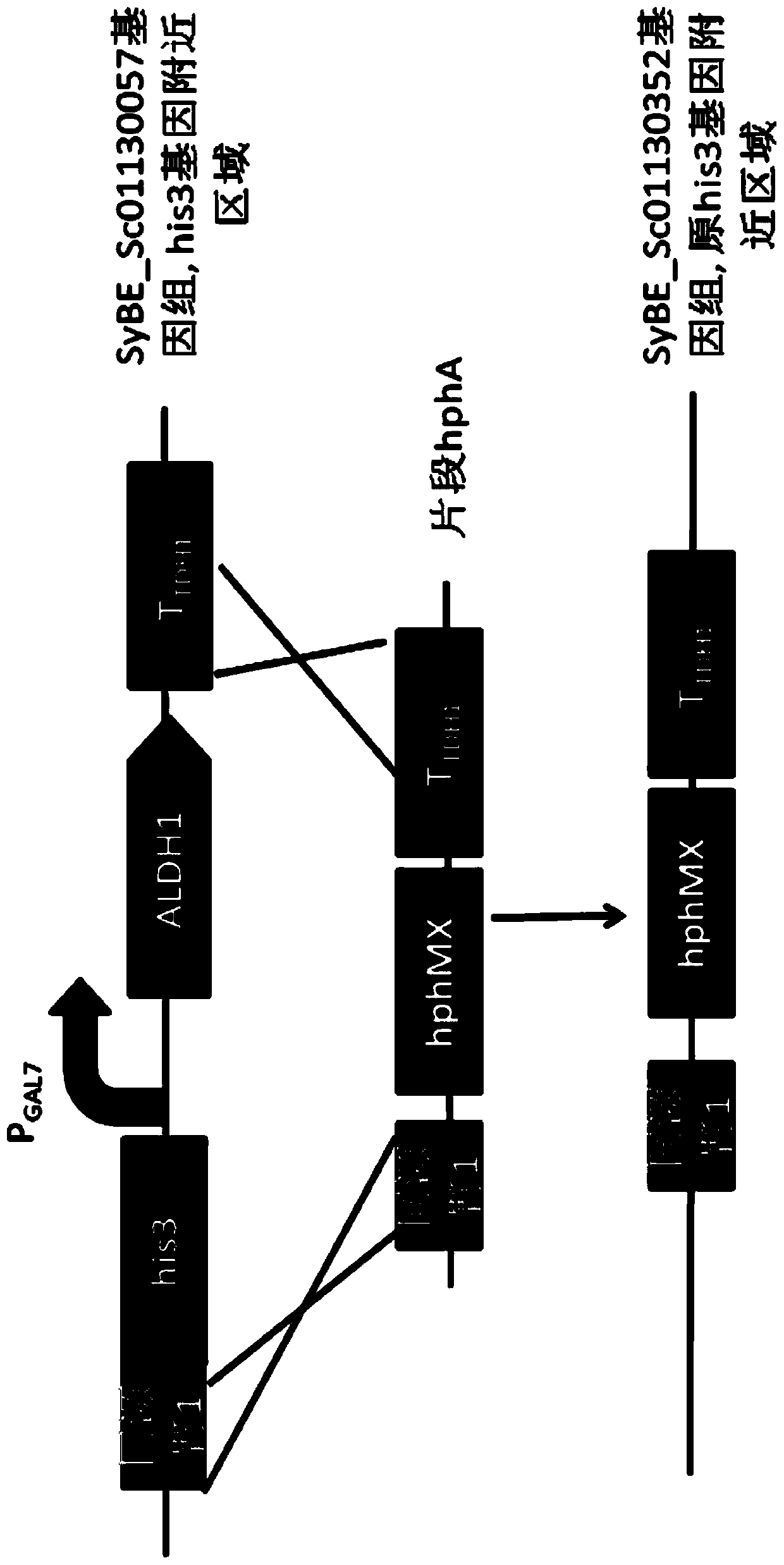
[0066] Example 1: Construction and import of module hphA
[0067] Using the plasmid SyBE_Ec01130018 as a template, fragment H0 (hphMX coding sequence) was amplified with 17E0b-hphA-F and 17E0b-hphA-R as primers; Hup was added (the homology arm 1 near the upstream His3 of the original ALDH1 coding sequence was used as the upstream homology arm); using the genome of SyBE_Sc01130057 as a template, Hdown was amplified with primers 17E0down-F and 17E0down-R (the original ALDH1 T downstream of the coding sequence TDH1 , as the downstream homology arm); then use the OE-PCR method to amplify the three fragments of H0, Hup, and Hdown using 17E0up-F, 17E0down-R as primers to amplify the module hphA, that is, the homology arm 1 (upstream of the His3 tag Partial sequence)+hphMX+T TDH1 . hphA was introduced into the strain SyBE_Sc01130057, and the strain SyBE_Sc01130352 was obtained through homologous recombination in yeast; the construction diagram will be figure 2 , see the import d...
Embodiment 2
[0068] Example 2: His3 tag (containing homology arm 1, that is, using the partial sequence upstream of the His3 tag as homology arm 1)+P GAL7 +ALDH1 H194R / ALDH1 V247F The coding sequence of +T TDH1 The construction and import of the module
[0069] 1. His3 tag +P GAL7 +ALDH1 V247F The coding sequence of +T TDH1 The construction and import of the module
[0070] The plasmid pSB1C3 was treated with EcoRI and PstI to obtain the fragment Vector-EP; using the genome of the bacterial strain SyBE_Sc01130057 as a template, the fragment E-His3, namely the His3 tag, was amplified by PCR with primers 17E1-His3-FE and 18E-His3-R;
[0071] Using SyBE_Ec01130018 as a template, the fragment E-ALDcF-a, namely P GAL7 +ALDH1 V247F The upper half of the coding sequence; using primers 18E5cF-F and 17E1-ALDH1-RP, the fragment E-ALDcF-b, namely ALDH1, was amplified by PCR V247F The lower half of the coding sequence;
[0072] The fragment E-ALDcF-a and fragment E-ALDcF-b were connected by...
Embodiment 3
[0083] Example 3: His3 tag (containing homology arm 1, that is, using the partial sequence upstream of the His3 tag as homology arm 1)+P GAL7 +DBR2-ALDH1 H194R The coding sequence of +T TDH1 The construction and import of the module
[0084] Treat the plasmid pSB1C3 with EcoRI and PstI to obtain the fragment Vector-EP; use the plasmid pRS423 as a template, and use primers 17E1-His3-FE and 17E1-His3-R to amplify the fragment E-His3, which is the His3 tag, by PCR;
[0085] Using SyBE_Ec01130021 as a template, the fragment E-DBR2, namely P GAL7 + DBR2 coding sequence;
[0086] Using SyBE_Ec01130018 as a template, using primers 17E1z-ALDH1-F and 18E5dR-R, the fragment E-ALD-1, ALDH1 was amplified by PCR H194R The lower half of the coding sequence; with SyBE_Ec01130018 as a template, using primers 18E5dR-F and 17E1-ALDH1-RP, the fragment E-ALD-2, ALDH1 was amplified by PCR H194R The upper half of the coding sequence;
[0087] Fragment E-His3 and fragment E-DBR2 were connected...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com