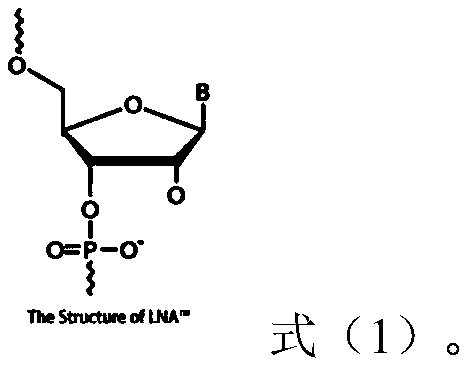
Kit for detecting mutation of locus L858R of EGFR gene
A site mutation and kit technology, applied in the field of nucleic acid detection, can solve problems such as low sensitivity and difficult DNA content, achieve high sensitivity, optimize the reaction system, and reduce human interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Design and synthesis of primer pairs and probes for detection of EGFR gene L858R site mutation
[0055] According to the 858th mutation of exon 21 of EGFR (i.e. L858R mutation, c2573T>G), design and synthesize the primer pair A and fluorescent probe B for detecting the L858R site mutation of the EGFR gene based on digital PCR technology; the specific sequence as follows:
[0056] The primer pair A includes an upstream primer and a downstream primer;
[0057] The nucleotide sequence of the upstream primer is: 5'-CACCGCAGCATGTCAAGATCA-3';
[0058] The nucleotide sequence of the downstream primer is: 5'-CTTTGCCTCCTTCTGCATGGTAT-3'
[0059] Fluorescent probe B includes mutant fluorescent probes and wild-type fluorescent probes;
[0060] The nucleotide sequence of the mutant fluorescent probe is: 5'-TTGGGCGGGCCAAAC-3';
[0061] The nucleotide sequence of the wild-type fluorescent probe is: 5'-TTGGGCTGGCCAAAC-3';
[0062] Both the 7th base at the 5' end of the ...
Embodiment 2
[0064] Example 2: A kit for detecting the L858R site mutation of the EGFR gene
[0065] The kit provided by the present invention includes: primer pair A provided in Example 1, fluorescent probe B, positive quality control substance, negative quality control substance and reaction premix.
[0066] The preparation method of the positive quality control product is as follows: artificially synthesize two kinds of DNA fragments with a length of 200 bp containing the wild-type and mutant-type mutation sites respectively, and load them into the plasmid vector pET-23d(+) (Promega). Use Qubit 3.0 for quantification, calculate the copy number concentration of the two types of plasmids, and mix the two plasmids (containing 1% L858R mutant DNA) according to the copy number ratio mutant type: wild type = 1:100, and then beat the plasmid mixture by ultrasound Break into fragments of about 180bp, quantify to 10ng / μL, and use it as a positive quality control.
[0067] The negative quality c...
Embodiment 3
[0070] Embodiment 3: the method for detection EGFR gene L858R site mutation
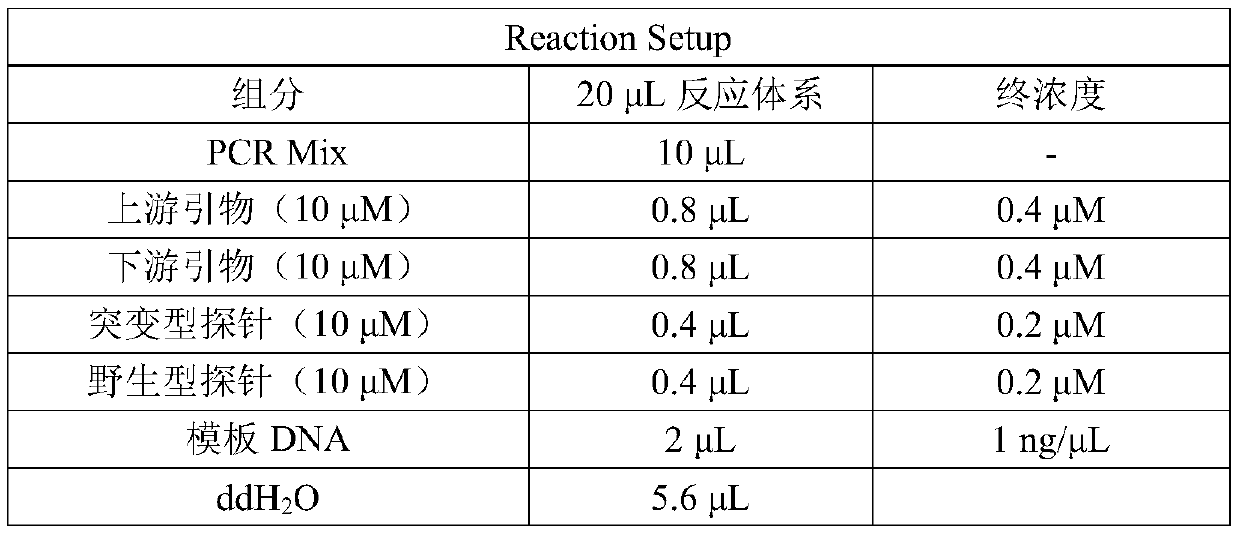
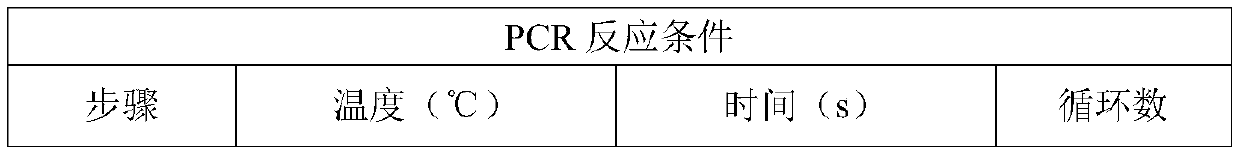
[0071] Using the kit described in Example 2, set up a PCR reaction system according to Table 1, and set up a PCR reaction system according to ddH 2 O. The order of the reaction master mix, probe, primer, and template DNA, add the above samples into the PCR tube according to the reaction system in Table 1, mix the mixed system for 15 seconds with a gentle vortex, and collect the solution by short-term centrifugation to the bottom of the test tube. Load the prepared reaction system onto the PCR chip to form a micro-reaction unit. Put the chip into a digital PCR instrument, perform PCR reaction according to the PCR reaction conditions in Table 2, and select FAM and CY3 as the channels for fluorescence detection.
[0072] Table 1: Reaction System
[0073]
[0074] Table 2: PCR reaction conditions
[0075]
[0076]
[0077] After the amplification is completed, the effective fluorescent posit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com