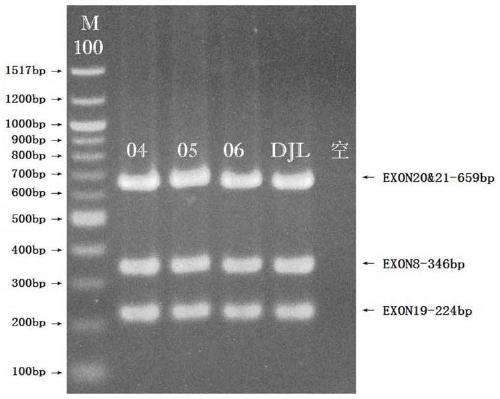
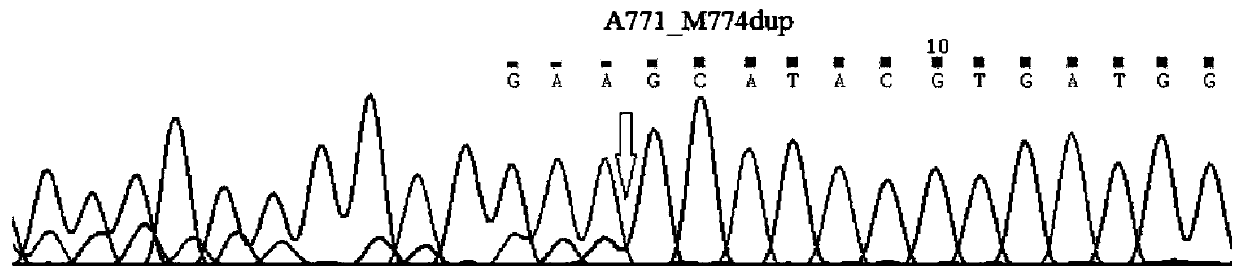
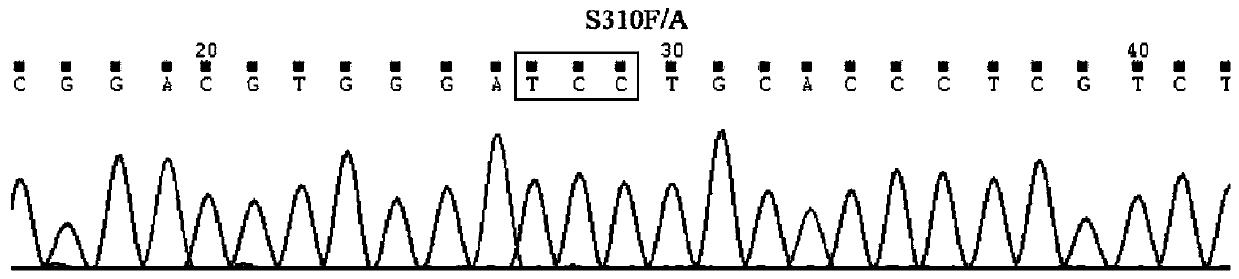
Method for synchronously detecting gene mutations of 8th, 19th, 20th and 21st exons of ERBB2 gene
A technology for simultaneous detection and detection methods, applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems of low detection throughput, improve detection throughput, save costs, The effect of reducing inspection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Design and synthesize primer sets, including the following steps:
[0036] Step 1.1: Based on exons 8, 19, 20, and 21 of the ERBB2 gene, design upstream and downstream primers and corresponding sequencing primers for specifically amplifying the mutation regions of exons 8, 19, 20, and 21 of the ERBB2 gene.
[0037] For the design of primers, Primer Quest and Primer Premier 5.0 were used to design primers and analyze dimers and stem-loop mismatches. Primers were designed at both ends containing mutation sites, and the annealing temperatures of the three pairs of primers were basically consistent.
[0038] The primer set provided in this example covers all gene mutations of exons 8, 19, 20, and 21 of the ERBB2 gene. Since small sequence changes will lead to a significant decrease in primer amplification efficiency and poor specificity, multiple PCR primer sets were designed for different sites / exons, and after pre-experimental screening, the length and position of the pro...
Embodiment 2
[0043] Genomic DNA, tumor genomic DNA or tumor cell-free DNA (ctDNA) is extracted from the sample to be tested, comprising the following steps:
[0044] Step 2.1: The samples to be tested are serum / plasma samples separated from fresh peripheral blood containing DNA, tumor tissue samples, oral exfoliated cells collected from oral swabs, or fresh peripheral blood samples.
[0045] In this embodiment, the sources of samples are tumor patients and normal human body.
[0046] Step 2.2: Use Qiagen QIAamp Circulating Nucleic Acid Kit (55114), or, use Tiangen Blood / Cell / Tissue Genomic DNA Extraction Kit (DP304), to extract tumor cell-free DNA (ctDNA), or, tumor For genomic DNA, use Tiangen oral swab genomic DNA extraction kit (DP322), or use blood / cell / tissue genomic DNA extraction kit (DP304) to extract genomic DNA from the sample to be tested, and use NP80-touch ( German IMPLEN) to measure the concentration and purity of DNA, and store the DNA.
Embodiment 3
[0048] A multiplex PCR detection method for synchronously detecting mutations in exons 8, 19, 20, and 21 of the ERBB2 gene, comprising the following steps:
[0049] Step 3.1: Use the DNA obtained in step 2.2 as an amplification template, and use the primer set synthesized in step 1.2 to configure a multiplex PCR reaction system.
[0050] In this example, the DNA polymerase and buffer in the KOD FX enzyme system (article number: KFX-101) of TOYOBO Co., Ltd. were used as the basic raw materials. concentration, buffer concentration, and enzyme dosage to prepare a multiplex PCR amplification system. The specific composition of this reaction system is shown in Table 2 below. Of course, the proportional amplification / reduction of the reaction system is within the protection scope of the embodiment of the present invention; the purpose of amplification can also be achieved by replacing other DNA polymerase systems and adjusting the appropriate ratio.
[0051] Table 2
[0052] ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap