RPA primer composition and method for detecting shiga toxin-producing Escherichia coli
An Escherichia coli and Shiga toxin-producing technology, applied in the biological field, can solve the problems of lack of degenerate bases, unclear genotype range, lack of detectable genotypes in test data, etc. Simple to use effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] This embodiment provides an RPA primer composition and probe for detecting Shiga toxin-producing Escherichia coli. The specific design process includes the following steps:
[0072] Download stx from the GeneBank database (www.ncbi.nlm.nih.gov / genbank)1 and stx 2 Nucleic acid sequence of genotype subtype, including stx 1 Three genotypes (stx 1a , stx 1c , stx 1d ) and stx 2 7 genotypes (stx 2a , stx 2b , stx 2c , stx 2d , stx 2e , stx 2f , stx 2g ). Primer Premier 5.0 software was used to design primers and probe combinations (Table 1) according to the RPA design principles of TwistDx Company, and were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0073] Table 1 RPA primer and probe sequences for detection of Shiga toxin-producing Escherichia coli
[0074]
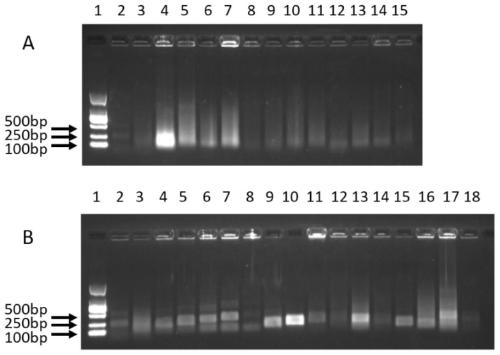
[0075] Use RPA technology to screen suitable RPA amplification primers. The specific operation steps are as follows:
[0076] (1) will detect stx 1 and stx 2 The ups...
Embodiment 2
[0082] This embodiment provides a method for detecting Shiga toxin-producing Escherichia coli, using fluorescent RPA technology; including the following steps:
[0083] In step 1, fluorescent RPA reaction uses fluorescent DNA constant temperature rapid amplification kit (Anpu Future Biotechnology Co., Ltd.). Add 2.95 μL of reaction premix A to the dry powder reaction tube, 0.1 μL each of the primers screened in Example 1 and the probe (10 μmol / L) provided, 1.5 μL of DNA template, and finally add buffer B (containing 280 mmol / L Mg 2+ ) 0.25 μL to make a 5 μL reaction system.
[0084] Step 2: Mix well, set at 39°C (temperature range: 37-42°C) for constant temperature reaction for 20min, and collect fluorescence signal every 30s for a total of 40 cycles.
[0085] Step 3: After the reaction is completed, the data is read and analyzed.
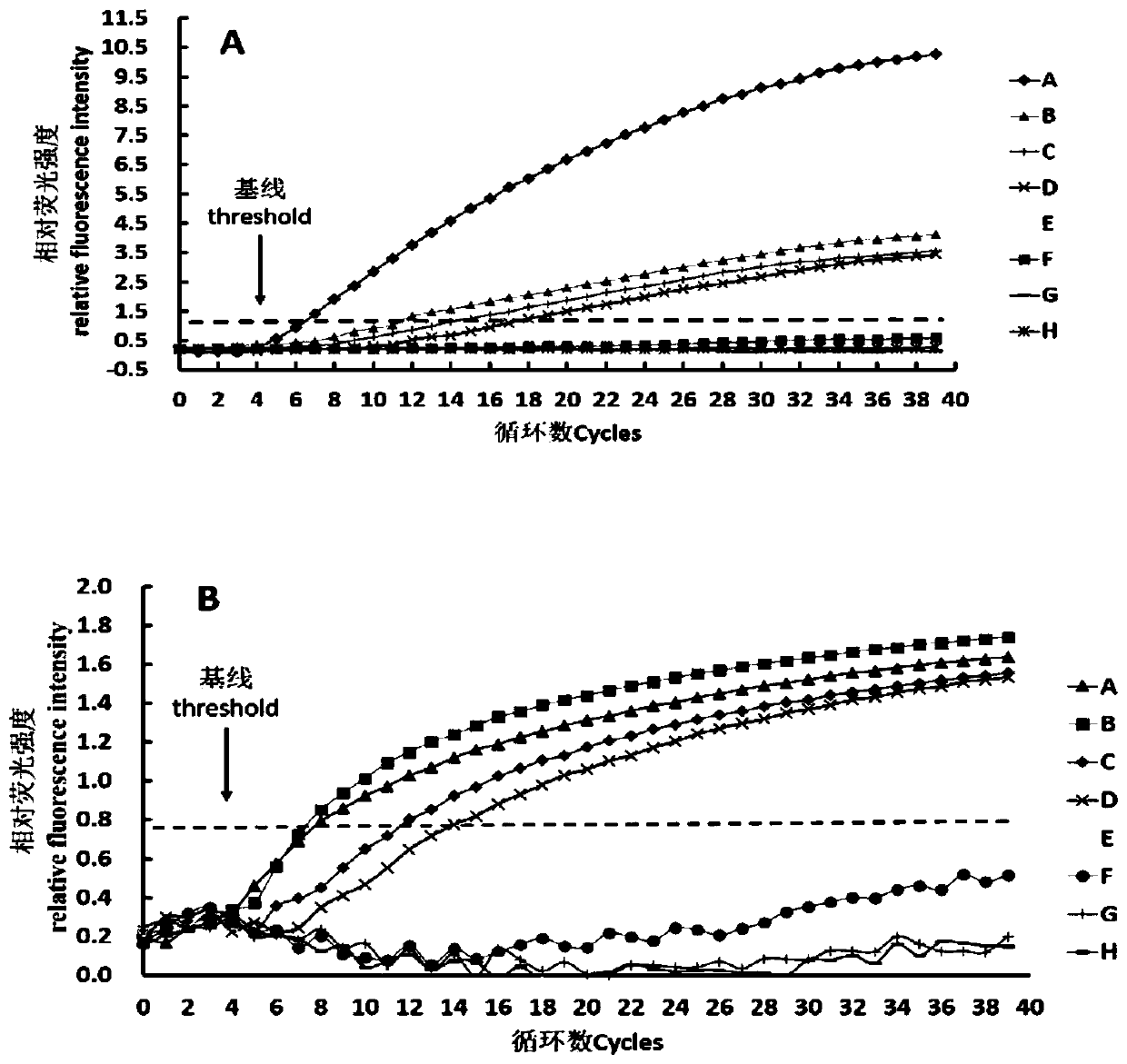
[0086] By detecting genomic nucleic acid and bacterial cells, the sensitivity of the method using fluorescent RPA technology provided by this em...
Embodiment 3
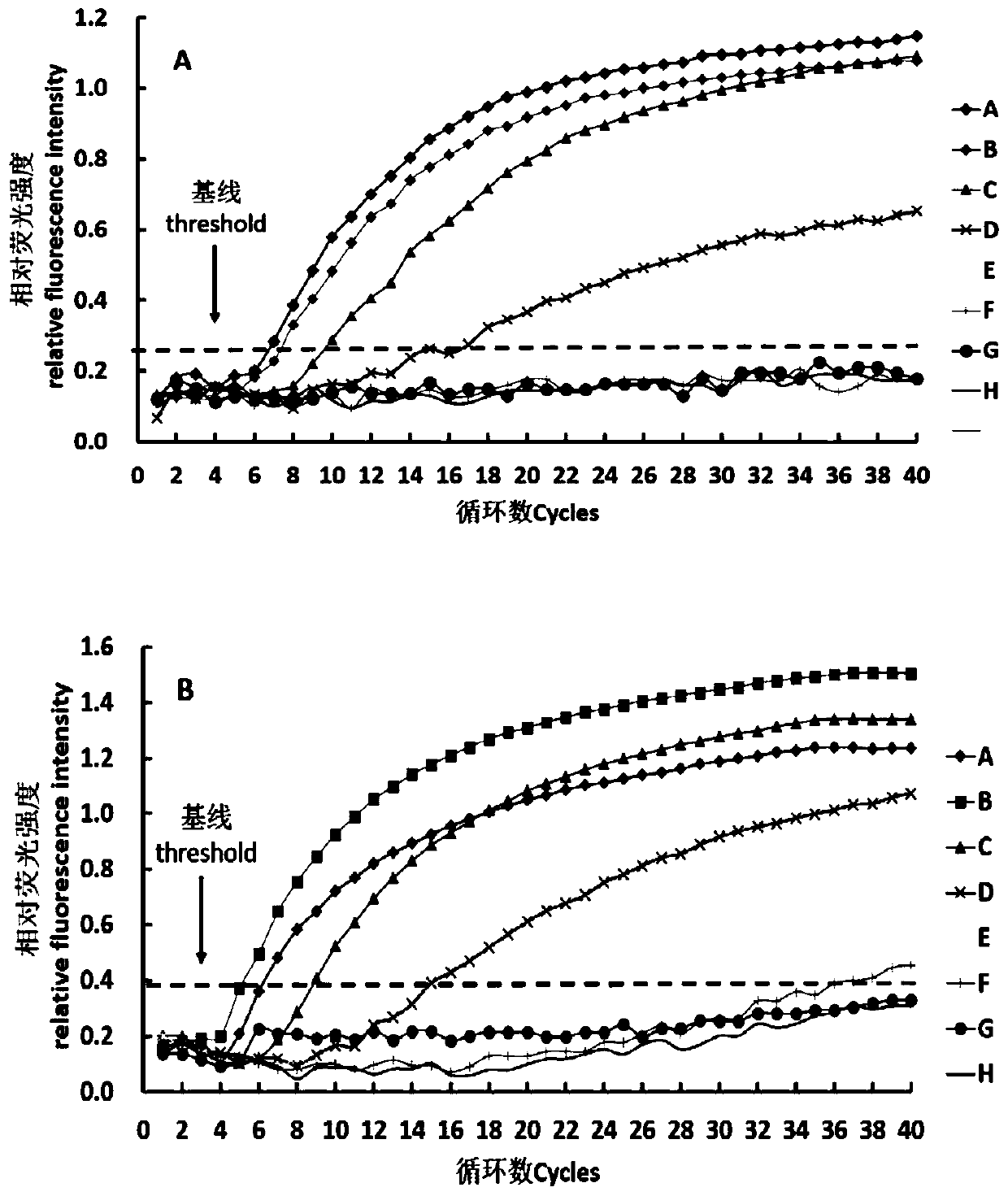
[0092] This example provides a method for detecting Shiga toxin-producing Escherichia coli, using fluorescent RPA microfluidic chip technology to simultaneously detect 9 gene subtypes of stx1a, stx1c, stx1d, stx2a, stx2b, stx2c, stx2d, stx2e and stx2g .
[0093] Among them, the disk centrifugal microfluidic chip assembly and chip detector (SX-MA2000 plus) used in the experiment were provided by Shanghai Suxin Biotechnology Co., Ltd. The chip is made of polycarbonate material and micro-injected into a disc structure. Each chip has 8 sets of identical microfluidic structure areas, and each area has 1 sample loading area, 1 liquid distribution area and 4 independent reactions. area, 32 reactions can be carried out simultaneously.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com