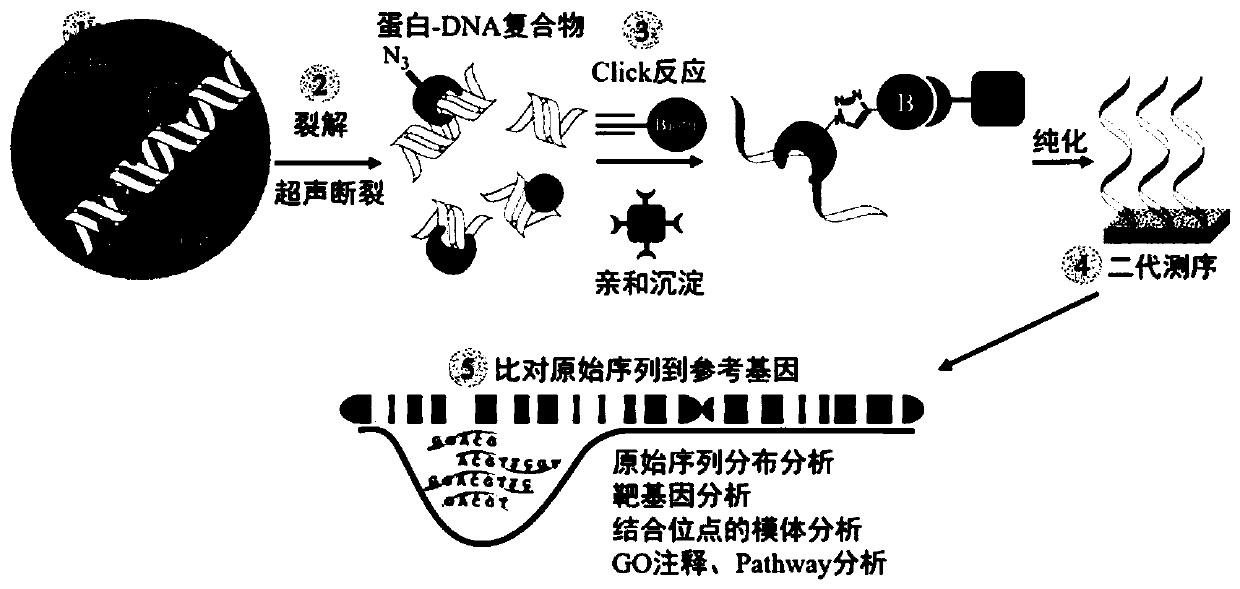
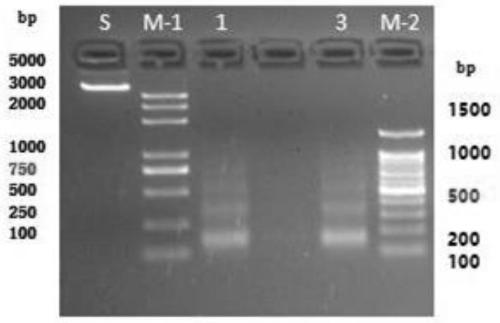
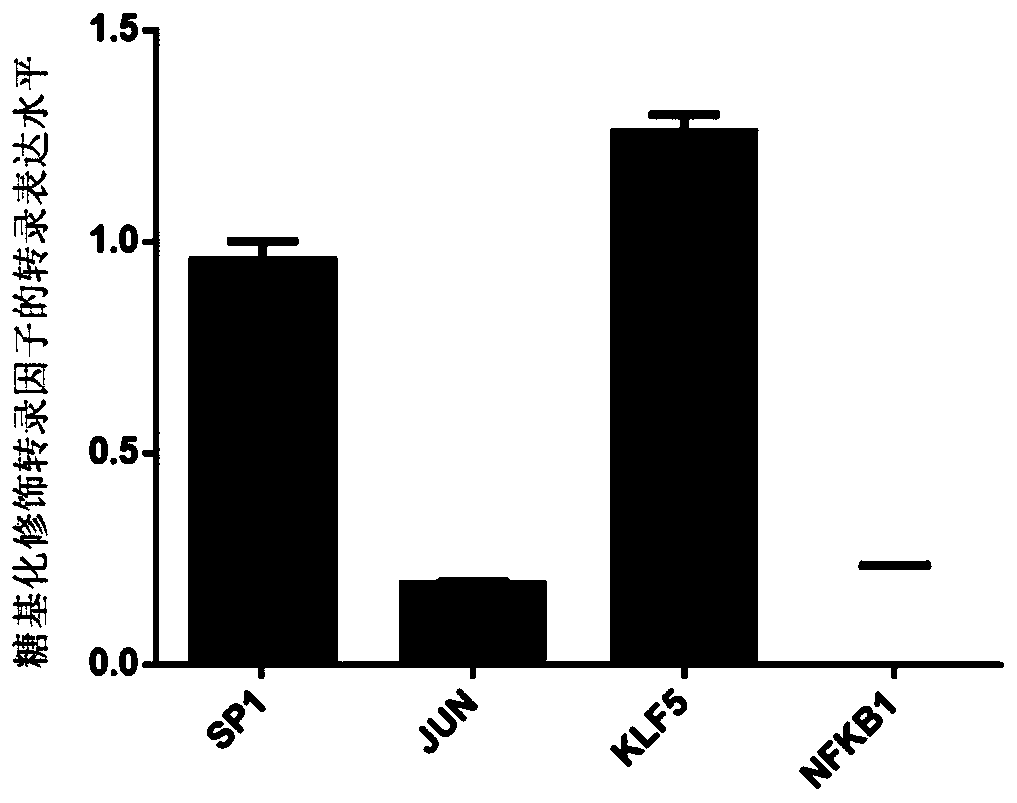
Method for obtaining O-GlcNAc modified transcription factor binding chromatin DNA sequence based on glycometabolism labeling
A DNA sequence and transcription factor technology, applied in the field of glycobiochemical chromatin immunoprecipitation, can solve the problems of inability to study glycotranscription factors and target gene DNA sequences, and achieve strong biological stability, high chemical selectivity, and good The effect of compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: This example provides a method for obtaining the O-GlcNAc glucose metabolism marker transcription factor group binding chromatin DNA sequence in MCF-7 cells.
[0025] The experiment includes the following steps:
[0026] S1. Cell labeling and cross-linking
[0027] MCF-7 cells were treated with 1mM GalNAz for 48h. Take out a plate of 10 cm dish cells and add formaldehyde for cross-linking, so that the final concentration of formaldehyde is 1%, incubate at room temperature for 10 min, add glycine with a final concentration of 0.125M to terminate cross-linking.
[0028] S2. Nuclei Preparation and Chromatin Digestion
[0029] Add 1 ml of ice-cold lysate containing 0.5 μM DTT and protease inhibitors to the cross-linked cells in S1. Incubate on ice for 10 min. Invert the tube every 3 minutes to mix. Nuclei were then pelleted by centrifugation. Resuspend the pellet in 1 ml ice-cold lysis buffer containing DTT. Resuspend in 100 μL of ice-cold lysate containin...
Embodiment 2
[0045] Example 2: This example provides a method for obtaining the O-GlcNAc glucose metabolism marker transcription factor group binding chromatin DNA sequence in Hela cells.
[0046] The experiment includes the following steps:
[0047] S1. Cell labeling and cross-linking
[0048] Hela cells were treated with 200μM Ac4GalNAz for 48h. Take out a plate of 10 cm dish cells and add formaldehyde for cross-linking, so that the final concentration of formaldehyde is 1%, incubate at room temperature for 15 minutes, add glycine with a final concentration of 0.125M to terminate cross-linking.
[0049] S2. Nuclei Preparation and Chromatin Digestion
[0050] Add 1 ml of ice-cold lysate containing 0.5 μM DTT and protease inhibitors to the cross-linked cells in S1. Incubate on ice for 10 min. Invert the tube every 3 minutes to mix. Nuclei were then pelleted by centrifugation. Resuspend the pellet in 1 ml ice-cold lysis buffer containing DTT. Resuspend in 100 μL of ice-cold lysate co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com