Application of USP33 as drug target in preparation of drugs
A technology for preparing drugs and drugs, applied in the field of protein engineering, can solve problems such as reducing the survival rate of Drosophila
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
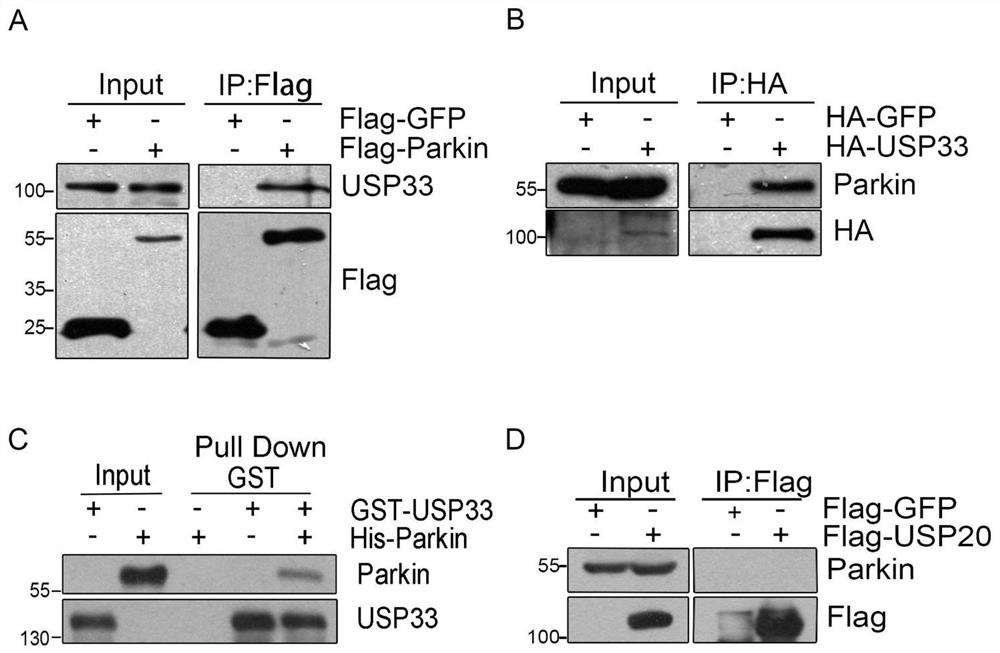
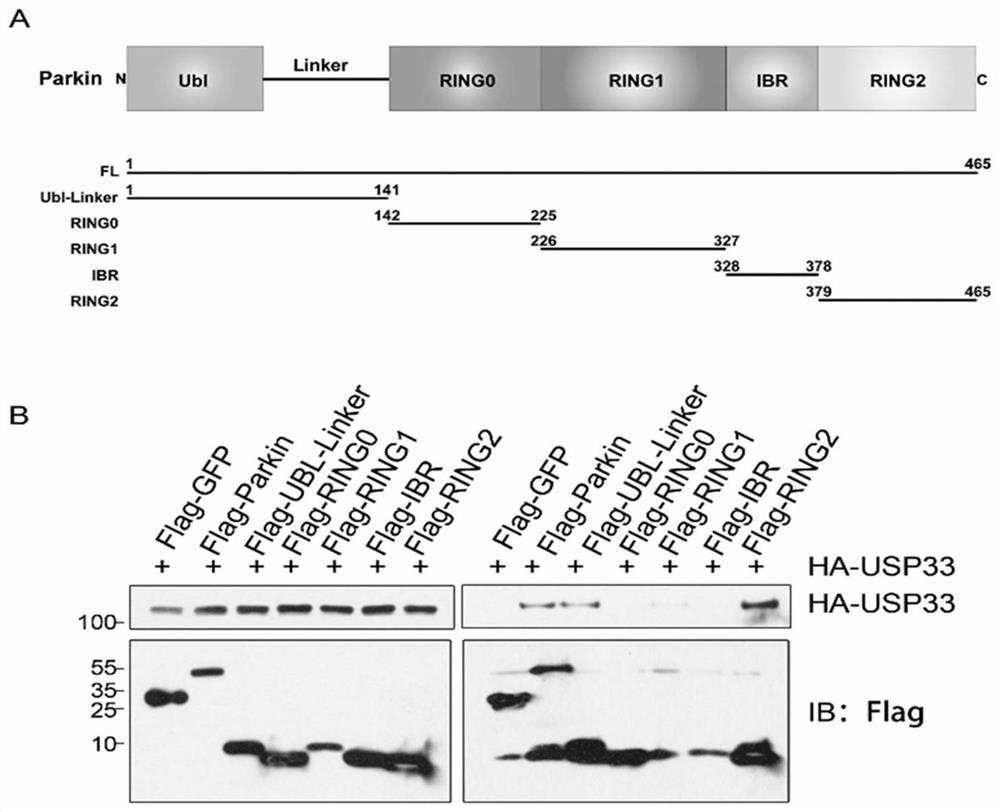
[0096] Example 1 Co-immunoprecipitation and mass spectrometry identification of the interaction between Parkin and the deubiquitinating enzyme USP33
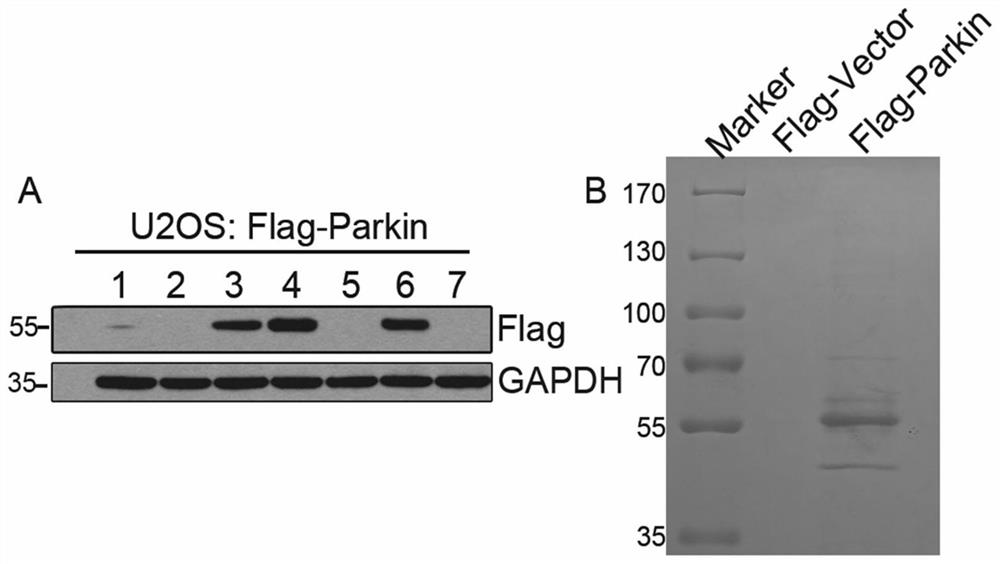
[0097] 1. The present invention utilizes co-immunoprecipitation combined with mass spectrometry to study Parkin's interacting protein network. In U2OS cells, the pcDNA3-Flag-Parkin plasmid was transfected, and the cells were screened by 400ug / ml G418 to obtain a cell line stably expressing the Flag-Parkin protein SEQ ID NO.2 ( figure 1 Figure A).
[0098] Cell No. 4 with a high expression level was selected for expansion culture, the whole cell lysate was collected, and anti-FlagBeads were incubated with it, the Flag-Parkin interacting protein was enriched by co-immunoprecipitation, and SDS-PAGE gel electrophoresis was used Separation of enriched protein samples ( figure 1 Figure B), and then stained with Coomassie Brilliant Blue G250 for protein profile analysis. The control group was transfected with pcDNA3-Flag-vector plas...
Embodiment 2
[0105] Example 2 Mitochondrial localization of USP33 protein
[0106] 1. USP33 is located in the mitochondrial outer membrane
[0107] The localization of endogenous USP33 in cells was verified by immunofluorescence experiments. In U2OS cells, mitochondria were labeled with Tom20 (mitochondrial outer membrane protein) antibody, and U2OS cells were labeled with USP33 antibody, as Figure 5 As shown in Figure A, it was found that endogenous USP33 and mitochondrial protein TOM20 had obvious co-localization, indicating that USP33 protein was located on mitochondria. Next, cell fractionation was performed on HEK293 cells, as Figure 5As shown in Figure B, the contamination of nuclear LaminB and cytoplasmic Tubulin was not detected in the mitochondrial fraction, indicating that the isolated mitochondria are of good purity and can be used in the next experiment. At the same time, Western blotting experiments confirmed that USP33 exists in mitochondria, as shown in the immunofluores...
Embodiment 3
[0111] Example 3 Knockdown of USP33 Promotes GFP-Parkin Translocation to Mitochondria
[0112] When cells are treated with CCCP (Carbonyl cyanide 3-chlorophenylhydrazone, inhibitor of mitochondrial oxidative phosphorylation), the mitochondria will be damaged and depolarized, and a large amount of Parkin protein will be recruited to the mitochondria to initiate the occurrence of mitophagy. In order to study whether the knockdown of USP33 will affect the process of mitophagy initiation, this example constructed a GFP-Parkin plasmid, transfected it into U2OS cells, and established a cell line stably expressing GFP-Parkin.
[0113] Cells stably expressing GFP-Parkin were treated with different concentrations of CCCP for 2 hours, and then the translocation of GFP-Parkin was observed, as shown in Figure 7 As shown, compared with the control group, GFP-Parkin aggregated and translocated in most of the cells treated at the concentration of 20 μM and 40 μM, while GFP-Parkin aggregated...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com