Molecular marker, detection primer and detection method for identifying lactobacillus helveticus, lactobacillus fermentum and lactobacillus acidophilus
A technology for Lactobacillus fermentum and Lactobacillus acidophilus, which is applied to the molecular markers of Lactobacillus fermentum and Lactobacillus acidophilus, the detection of primers, and the identification of Lactobacillus helveticus. It is difficult to popularize and apply problems in grassroots laboratories, and achieve the effects of short detection cycle, low detection cost and high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
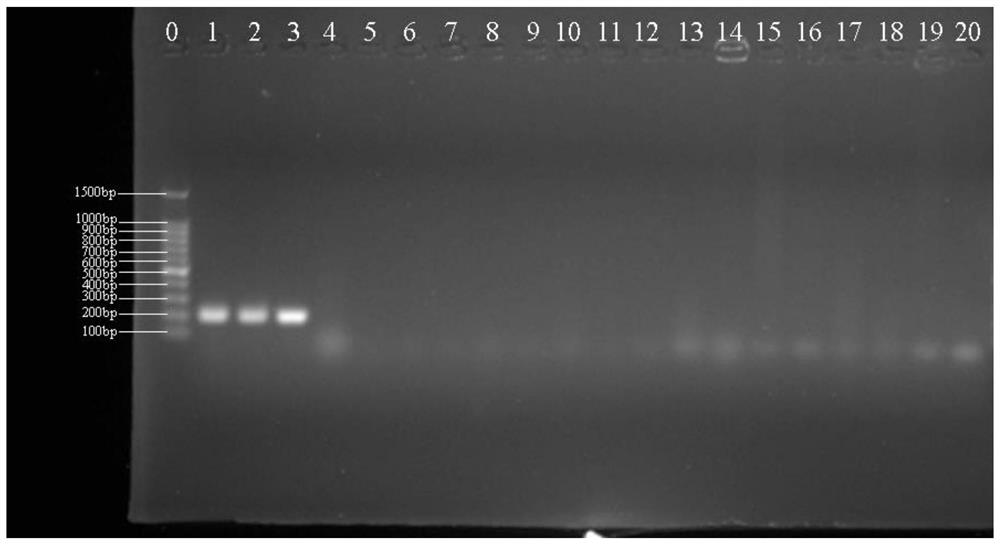
[0066] Embodiment 1: identification of Lactobacillus helveticus
[0067] 1. Bacterial DNA extraction
[0068] Take 1 mL of the bacterial culture and centrifuge at 12000r / min for 5min, discard the supernatant, collect the bacteria, add 50μL of the lysate of the bacterial DNA extraction kit, suspend and mix well, place in a water bath at 100°C for 15min, ice bath for 3min, and centrifuge at 12000r / min After 10 minutes, the supernatant was taken for later use, and the supernatant contained bacterial DNA as template DNA.
[0069] 2. PCR amplification
[0070] PCR uses a 25 μL reaction system, 1.0 μL of upstream and downstream target DNA-specific primers at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, and 25 mmol / L MgCl 2 2 μL, 2 μL of 10 mmol / L dNTP, 2.5 μL of 10×PCR buffer, 2.0 μL of DNA template, and 25 μL of sterile deionized water. PCR amplification conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 1 min, 58°C annealing for 45 s, 7...
Embodiment 2
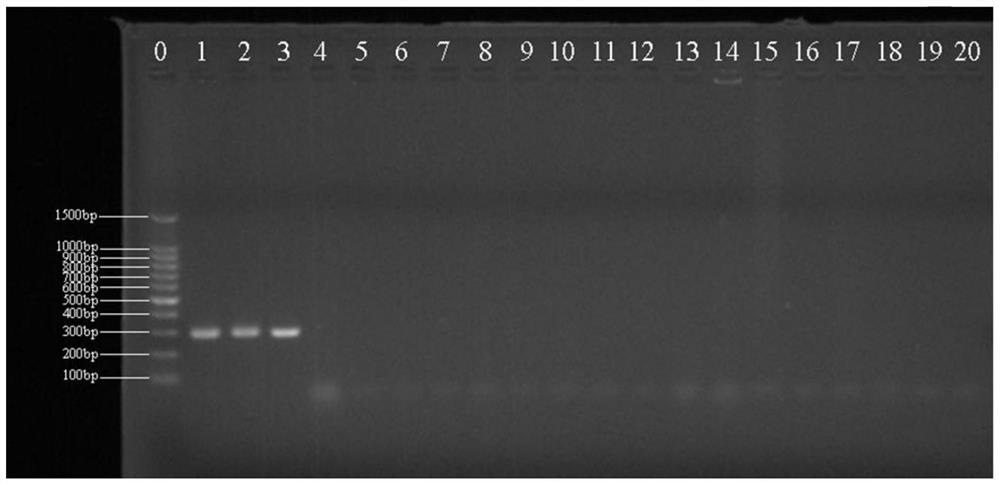
[0073] Embodiment 2: Identification of Lactobacillus fermentum
[0074] 1. Bacterial DNA extraction
[0075] Take 1 mL of the bacterial culture and centrifuge at 12000r / min for 5min, discard the supernatant, collect the bacteria, add 50μL of the lysate of the bacterial DNA extraction kit, suspend and mix well, place in a water bath at 100°C for 15min, ice bath for 3min, and centrifuge at 12000r / min After 10 minutes, the supernatant was taken for later use, and the supernatant contained bacterial DNA as template DNA.
[0076] 2. PCR amplification
[0077] PCR uses a 25 μL reaction system, 1.0 μL of upstream and downstream target DNA-specific primers at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, and 25 mmol / L MgCl 2 2 μL, 2 μL of 10 mmol / L dNTP, 2.5 μL of 10×PCR buffer, 2.0 μL of DNA template, and 25 μL of sterile deionized water. PCR amplification conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 1 min, 58°C annealing for 45 s, 72...
Embodiment 3
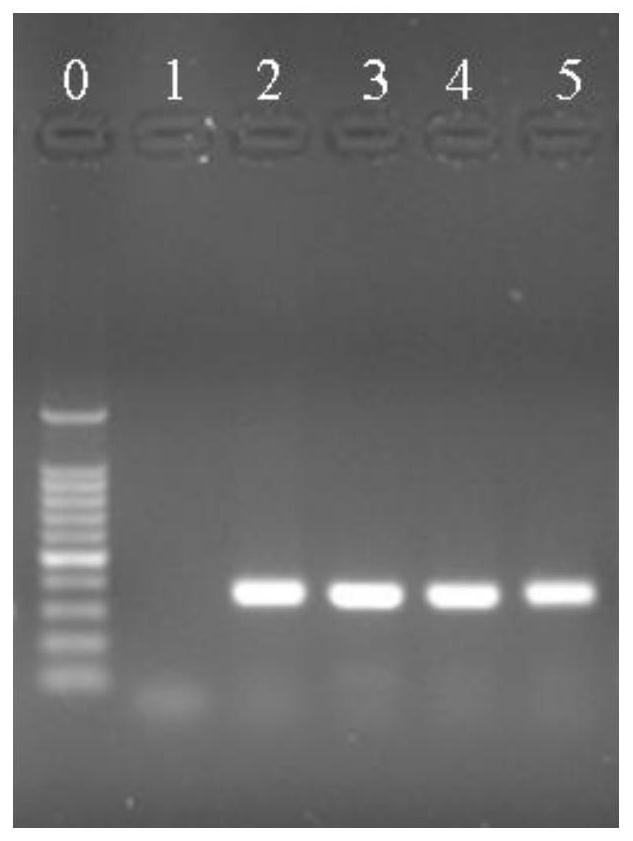
[0080] Example 3: Identification of Lactobacillus acidophilus
[0081] 1. Bacterial DNA extraction
[0082] Take 1 mL of the bacterial culture and centrifuge at 12000r / min for 5min, discard the supernatant, collect the bacteria, add 50μL of the lysate of the bacterial DNA extraction kit, suspend and mix well, place in a water bath at 100°C for 15min, ice bath for 3min, and centrifuge at 12000r / min After 10 minutes, the supernatant was taken for later use, and the supernatant contained bacterial DNA as template DNA.
[0083] 2. PCR amplification
[0084] PCR uses a 25 μL reaction system, 1.0 μL of upstream and downstream target DNA-specific primers at a concentration of 10 μmol / L, 0.5 μL of 1.5U Taq DNA polymerase, and 25 mmol / L MgCl 2 2 μL, 2 μL of 10 mmol / L dNTP, 2.5 μL of 10×PCR buffer, 2.0 μL of DNA template, and 25 μL of sterile deionized water. PCR amplification conditions were: 95°C pre-denaturation for 5 min; 95°C denaturation for 1 min, 58°C annealing for 45 s, 72°...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com