Genetically engineered bacterium for producing 2'-fucosyllactose and application of genetically engineered bacterium
A technology of fucosyllactose and engineering bacteria is applied in the fields of metabolic engineering and food fermentation, which can solve the problems of expensive glycoside donors and low synthesis yield, and achieve the effect of wide industrial application value.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1: Construction of the recombinant expression plasmid containing T7 promoter
[0050] The specific steps for constructing the recombinant expression vector are as follows (see Table 1 for the primer sequences involved):
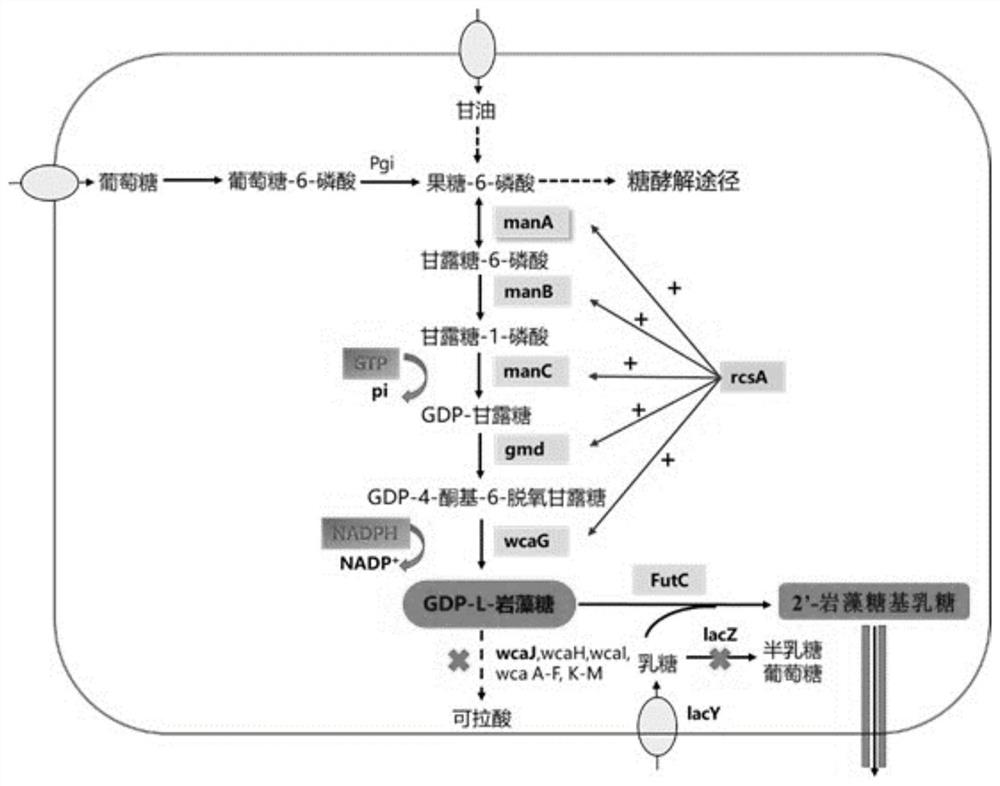
[0051] (1) Obtaining manB, manC, gmd and wcaG fragments: using the genome of Escherichia coli K12 as a template, using primers manB_F1 / R1, manC_F1 / R1, gmd_F1 / R1 and wcaG_F1 / R1 to perform PCR amplification, gel recovery DNA, and obtain manB, manC, gmd and wcaG gene fragments.
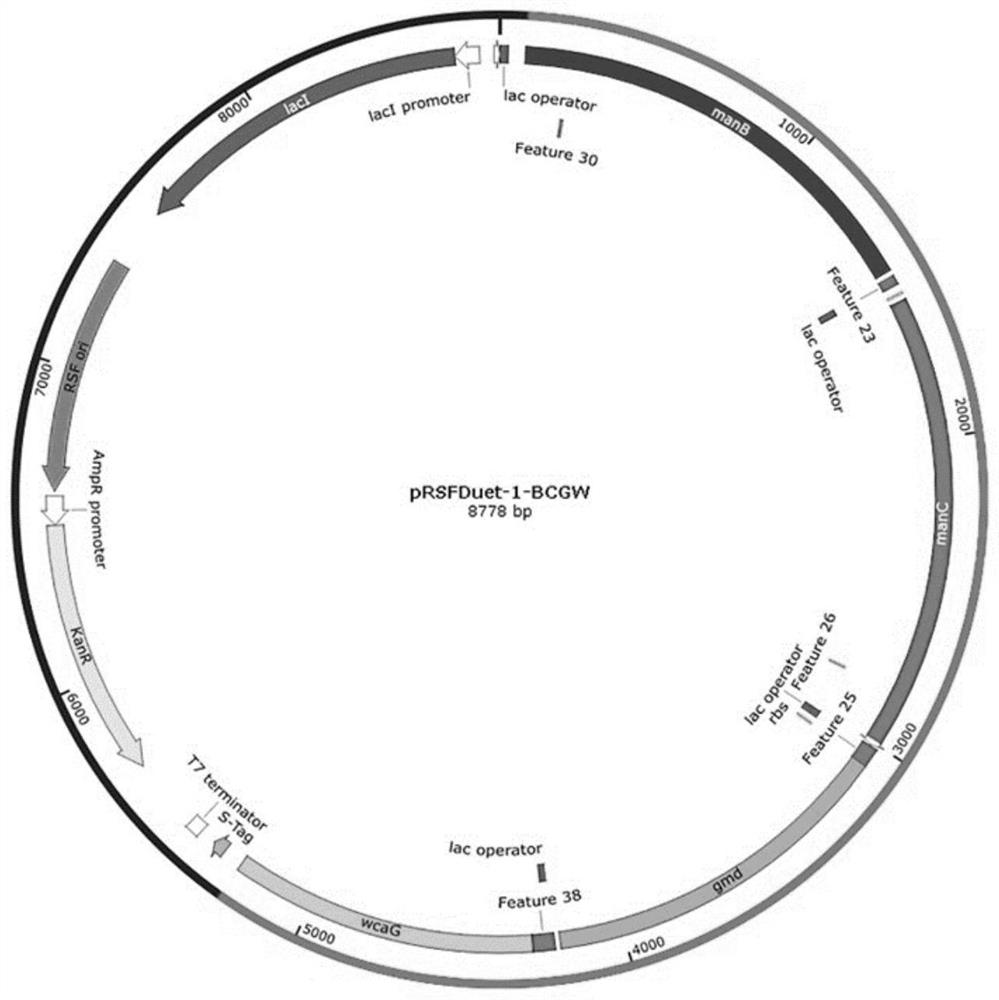
[0052] (2) Using pRSFDuet-1, pETDuet-1, pCDFDuet-1, pACYCDuet-1 and pCOLADuet-1 empty plasmids as templates, use V1_F / R to amplify the vector backbone. According to the In-Fusion cloning technology, the manB fragment was connected with pRSFDuet-1, pETDuet-1, pCDFDuet-1, pACYCDuet-1 and pCOLADuet-1 respectively, and positive clones were screened and sequenced to obtain recombinant plasmids pRSFDuet-1-manB, pETDuet- 1-manB, pCDFDuet-1-manB, pACYCDuet-1-manB, and pCOLAD...
Embodiment 2
[0061] Embodiment 2: Construction of the recombinant expression plasmid containing Trc promoter
[0062] The specific steps of recombinant expression vector construction are as follows (see Table 2 for the primer sequences involved):
[0063](1) Construction of pRSFDuet-Trc, pETDuet-Trc, pCDFDuet-Trc, pACYCDuet-Trc and pCOLADuet-Trc vectors: using pTrcHis2B as a template, using primers Trc_F / R to amplify the Trc promoter and MCS region, using pRSFDuet-1, The pETDuet-1, pCDFDuet-1, pACYCDuet-1 and pCOLADuet-1 empty plasmids were used as templates, and the vector backbone sequence was amplified using primers VDuet_F / R. According to the In-Fusion cloning technique, the two fragments were ligated and transformed into DH5α, positive clones were screened, and an expression vector containing a Trc promoter was finally obtained.
[0064] (2) Obtaining manB, manC, gmd and wcaG fragments: using the genome of Escherichia coli K12 as a template, using primers manB_F2 / R2, manC_F2 / R2, gmd_...
Embodiment 3
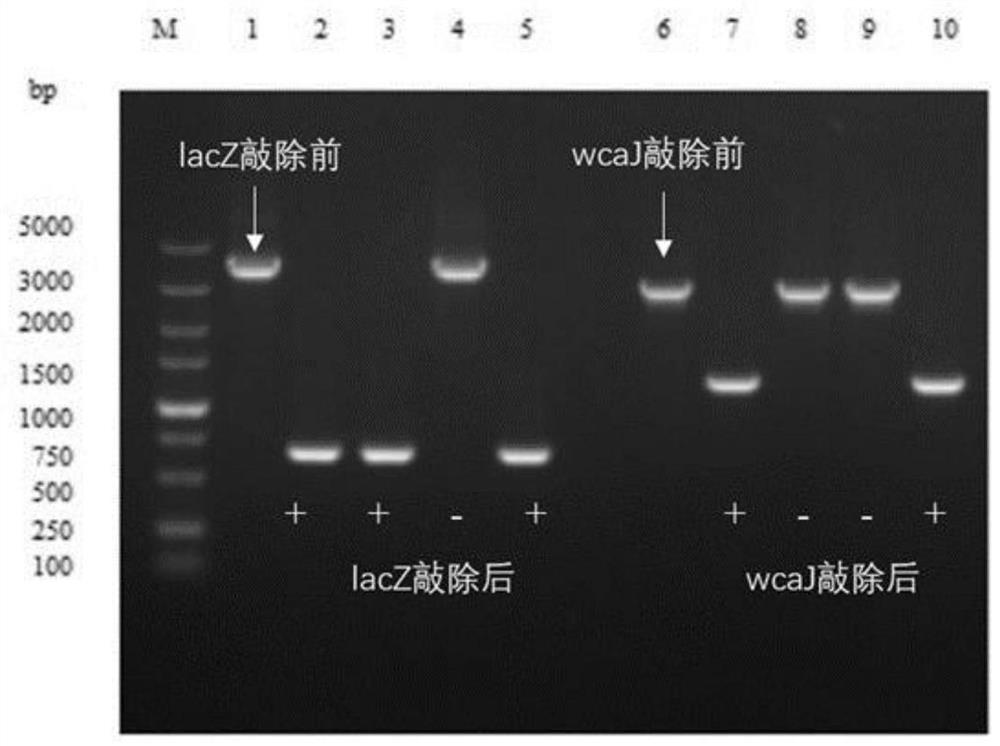
[0074] Example 3: Knockout of the lacZ and wcaJ genes of Escherichia coli BL21 (DE3) based on the CRISPR-Cas9 gene editing system
[0075] The specific steps of gene knockout are as follows (see Table 3 for the primer sequences involved):
[0076] (1) Taking the lacZ gene as an example, design the gRNA of the lacZ gene through http: / / www.regenome.net / cas-offinder / , introduce the designed 20bp sequence into gRNAR, use gRNAF / gRNAR as the upstream and downstream primers, and the pTargetF plasmid is template for PCR amplification. The amplified product was digested with the restriction endonuclease Dpn I to remove the redundant circular pTargetF plasmid. The amplified product was transformed into Escherichia coli DH5α competent cells, the plasmid was extracted, and identified by primers gRNAPF / gRNAPR sequencing, and the successfully constructed knockout plasmid was named pTF-ΔlacZ.
[0077] (2) Using the Escherichia coli BL21 (DE3) genome as a template, the upstream homology arm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com