Fluorescent biosensor for detecting mercury ions
A biosensor, mercury ion technology, applied in the field of biosensors, can solve the problems of convenience, speed, sensitivity, complex sample pretreatment, and high detection cost that are difficult to adapt to mercury ion detection, and achieve fast detection speed, low detection limit, Fast response time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
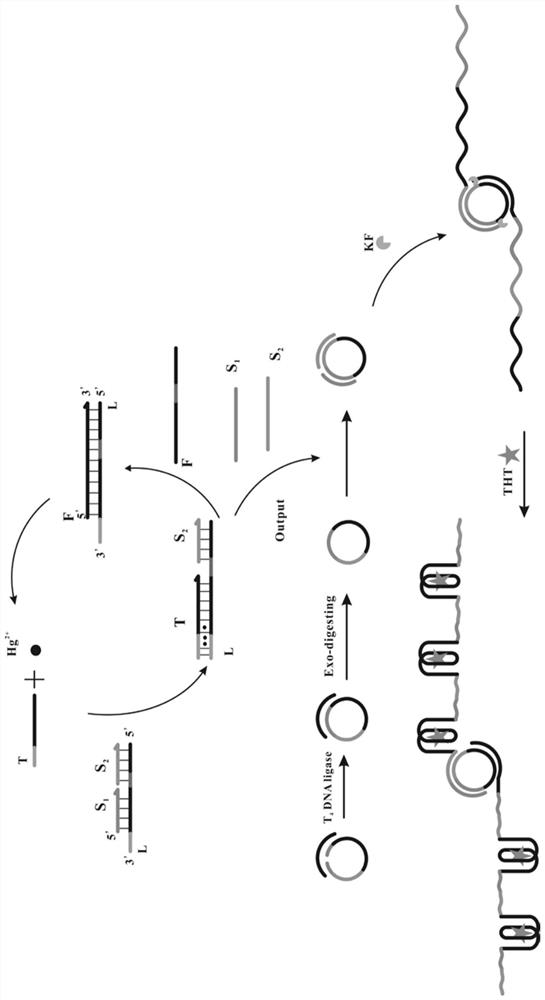
[0038] Example 1 Preparation of Composite Probe Q
[0039] (1) Synthesizing S1, S2 and L according to the sequences shown in SEQ ID No: 1-3;
[0040] (2) Dilute sterilized water (36 µL), 10×PBS buffer (pH7.4) (6 µL), S1 (6 µL, 100 µM), S2 (6 µL, 100 µM) and L (6 µL, 100 μM) was added to the pre-prepared sterilized EP tube, shaken for 20-30s, then incubated at 95°C for 5min, cooled slowly to room temperature and hybridized, and stored at -20°C for subsequent experiments.
Embodiment 2
[0041] Example 2 Preparation of cyclic template CP
[0042] (1) Synthesize 5' phosphorylated padlock probe TP and ligation probe LP according to the sequences shown in SEQ ID No: 5 and 6;
[0043] (2) Mix TP and LP with equal volume and molar concentration (6 µL, 100 µM), add 10×T4 DNA ligase buffer (6 µL) and incubate at 95°C for 5 min, cool to room temperature, then add T4 DNA Ligase (3 µL, 400 U / µL), incubated overnight at 16°C, then heated at 65°C for 10 min to inactivate the enzyme; add Exo I enzyme (2 µL, 20 U / µL) and Exo III enzyme (1 µL, 100 U / µL) was digested at 37°C for 2 h, then heated at 85°C for 20 min to inactivate the enzyme to obtain the circular template CP with the sequence shown in SEQ ID No: 4, and finally stored at 0-4°C for subsequent experiments .
Embodiment 3
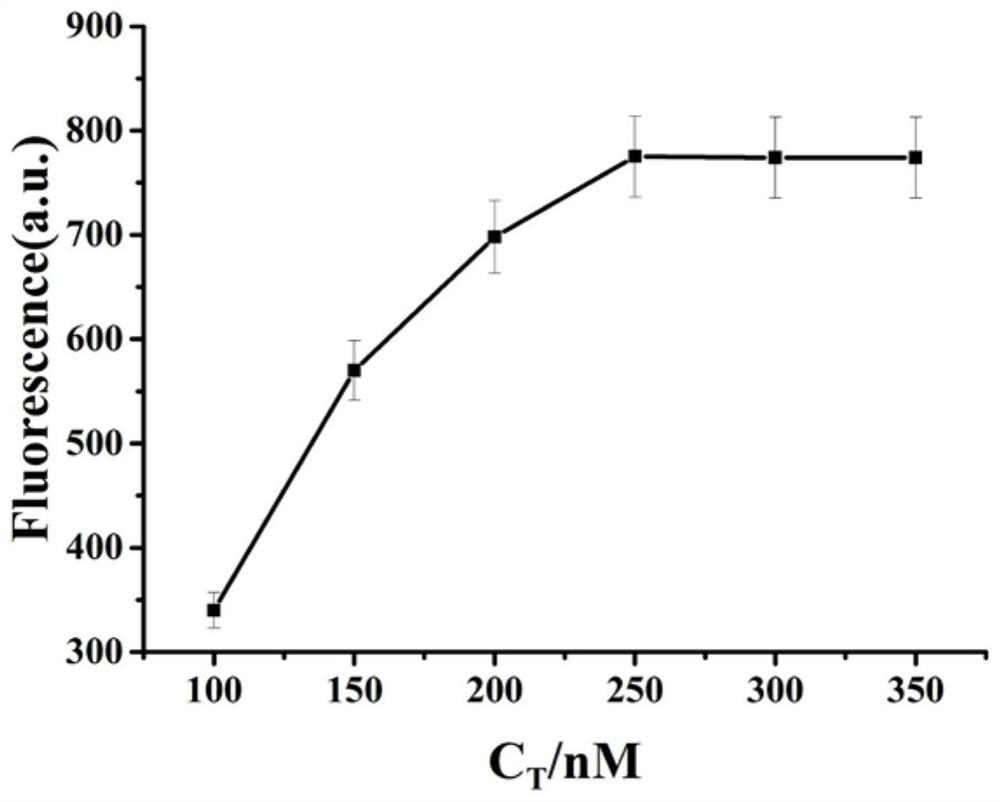
[0044] Example 3 Trigger chain T concentration screening
[0045](1) Dilute sterilized water (6 µL), 10× buffer 2 (3 µL), Hg 2+ (3 µL, 10 µM), trigger chain T (3 µL, make the final concentration 100nM, 150nM, 200nM, 250nM, 300nM, 350nM), composite probe Q (3 µL, the final concentration is 2 µM), fuel chain Add F (3 µL), circular template CP (3 µL, 1 µM), KF (3 µL, final concentration 0.5 U / µL) and ThT (3 µL, final concentration 10 µM) into the centrifuge tube, shake 20-30s, in a water bath at 37°C for 2h, the product was obtained;
[0046] (2) Dilute the above product to 100 µL, and measure the fluorescence intensity at 485 nm with 425 nm as the excitation wavelength;
[0047] see results figure 2 , it can be seen from the figure that with the increase of the concentration of the trigger chain T, the fluorescence intensity obtained in the experiment is continuously enhanced. is 250nM.
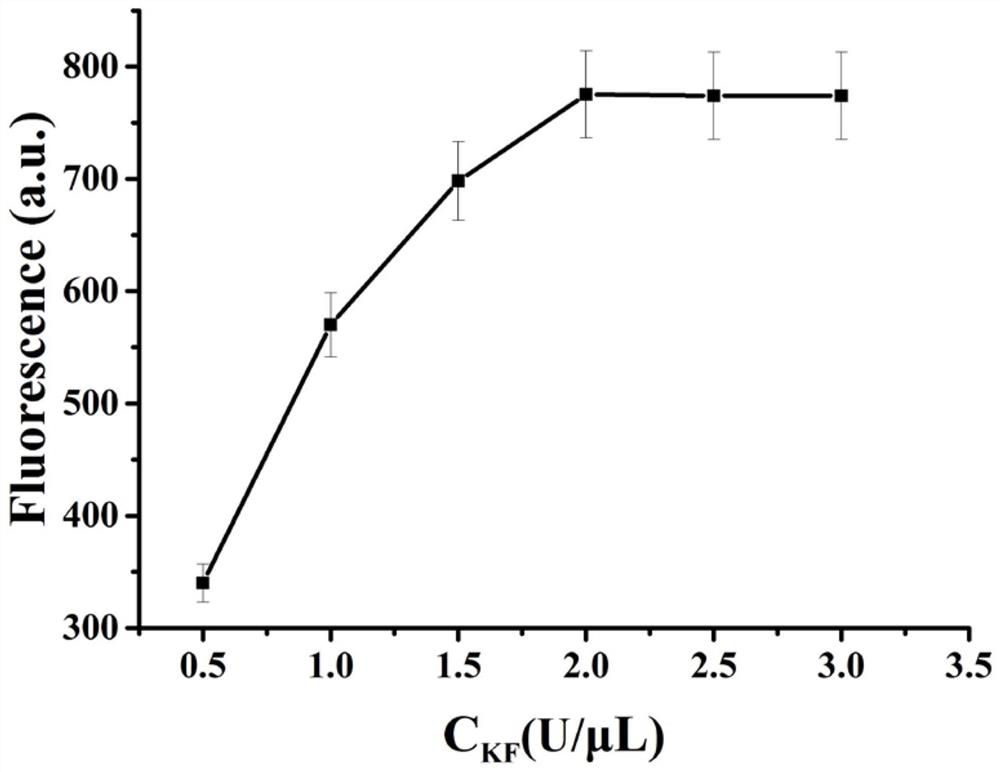
[0048] Example 3 KF concentration screening
[0049] (1) Dilute sterilized water (6 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com