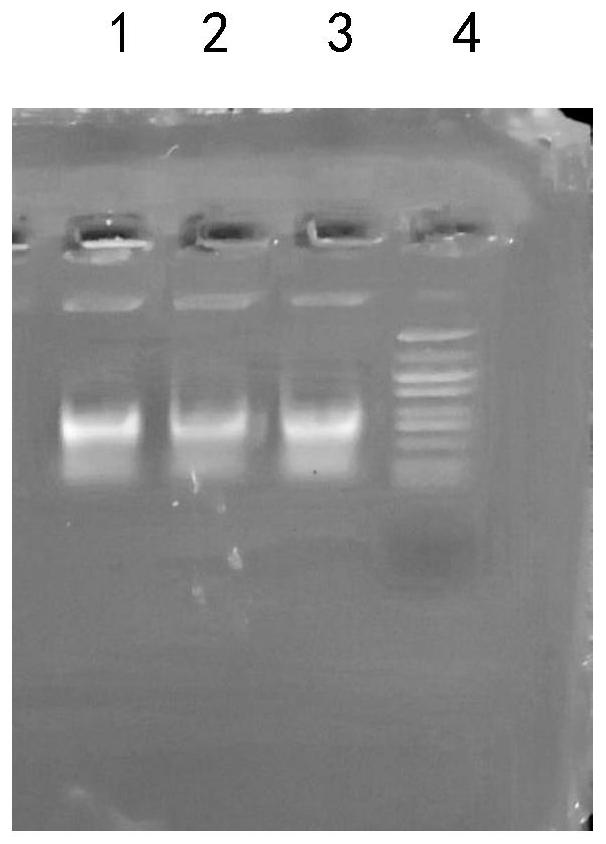
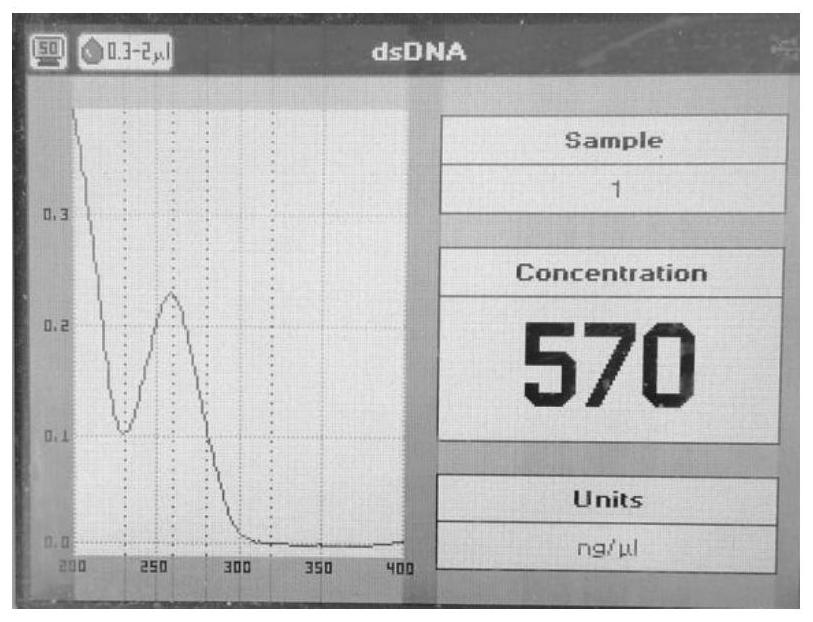
DNA extracting solution and method for extracting DNA of gram-positive bacteria
A technology of gram-positive bacteria and extract solution, which is applied in the field of DNA extraction and DNA extract solution of gram-positive bacteria, and can solve problems such as health threats to operators, PCR amplification, poor quality, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The extraction of embodiment 1 escherichia coli genomic DNA
[0031] Sufficiently resuspend the Escherichia coli cells in 2 ml of culture with 1 ml of sterile distilled water, centrifuge at 10,000 rpm for 1 min, discard the supernatant to collect the cells, and use solution I (chlorinated Sodium 0.15mol / L, EDTA 10mmol / L, Tris-HCl 15mmol / L with a pH value of 8.5) 450 μL resuspended, standing at 37°C to lyse the cell wall; ethanolamine) 50 μL and mix them upside down, and let stand at room temperature for 10 minutes; add 250 μL of solution III (3mol / L potassium chloride) in an ice bath, centrifuge at 10,000 rpm for 4 minutes after mixing, take the supernatant and transfer it to another test tube, add salicylic acid Methyl ester 500μL, vortex and mix until milky, centrifuge at 11000rpm for 2min, take the upper aqueous phase into another test tube (be careful not to absorb the protein layer and organic phase below), add an equal volume of isopropanol to precipitate for 5min...
Embodiment 2
[0033] The extraction of embodiment 2 Staphylococcus genomic DNA
[0034] Sufficiently resuspend the Staphylococcus cells in 2 ml of the culture with 1 ml of sterile distilled water, centrifuge at 10,000 rpm for 1 min, discard the supernatant to collect the cells, and use solution I (sodium chloride) containing 5 mg / ml lysozyme (added just before use) 0.2mol / L, EDTA20mmol / L, Tris-HCl 20mmol / L with a pH value of 8.5) 450μL resuspended, standing at 37°C to lyse the cell wall; ) 50 μL and mix them upside down, and let stand at room temperature for 12 minutes; add 250 μL of solution III (2.5mol / L potassium chloride) in an ice bath, centrifuge at 11,000 rpm for 3 minutes after mixing, take the supernatant and transfer it to another test tube, add salicylic acid Methyl ester 500μL, vortex and mix until milky, centrifuge at 11000rpm for 2min, take the upper aqueous phase into another test tube (be careful not to absorb the protein layer and organic phase below), add an equal volume o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com