Method for detecting flavoprotein based on three-dimensional fluorescence spectrum
A three-dimensional fluorescence, flavoprotein technology, applied in the field of enzyme protein detection, can solve the problems affecting the accurate, rapid and high-sensitivity of flavoprotein to characterize the target protein, and achieve the effects of high accuracy, high sensitivity and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1. Utilize the method of three-dimensional fluorescence spectrum to detect flavoprotein
[0040] Detection of sarcosine oxidase (SOX enzyme)
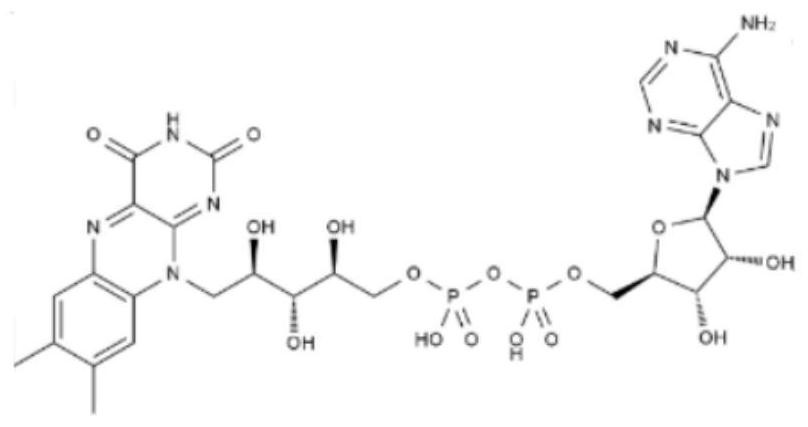
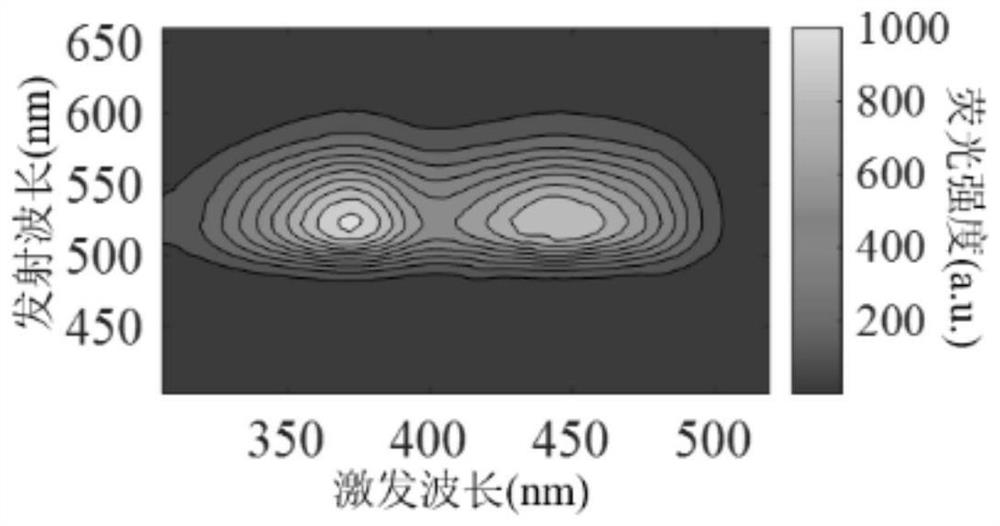
[0041] 1. Prepare 5 μmol FAD solution, store it away from light, take 3mL FAD solution, and use the F-7000 fluorescence spectrometer to collect the FAD spectrum under the condition of PMT voltage of 700V. image 3 As shown, the results show that the FAD fluorescence characteristic peaks are in the range of λex=330-500nm and λem=500-600nm, and the maximum fluorescence intensity of the FAD fluorescence peak appears at λex / λem=396 / 456nm.
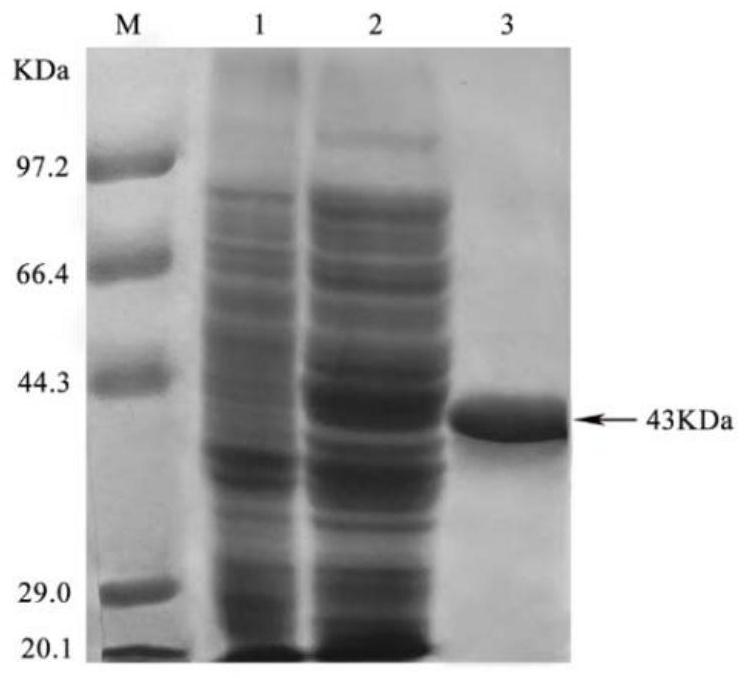
[0042] 2. Preparation of recombinant sarcosine oxidase (SOX) pure enzyme
[0043] The sarcosine oxidase gene (GenBank: AY626822.2) derived from Bacillus sp.BSD-8 was cloned into the vector pET28a, and the recombinant expression plasmid pET28a-sox was transformed into E.coli BL21(DE3) for inducible expression. Pick a single colony from the solid plate, insert it into LB medium, and cul...
Embodiment 2
[0054] Example 2. Using three-dimensional fluorescence spectroscopy to quantify sarcosine oxidase
[0055] 1. Determination of the position of the endogenous fluorescence characteristic peak of pure sarcosine oxidase
[0056] Sarcosine oxidase pure enzyme liquid samples with different protein concentrations were selected according to the gradient to measure the FAD fluorescence intensity at 700PMT voltage, and the sarcosine oxidase protein concentration (c, mg / mL) was compared with the FAD fluorescence intensity (ΔF, a.u.) Linear fitting, the concentration of SOX active protein has a good linear relationship in the range of 0.05-1.0 mg / mL, according to the linear relationship between the fluorescence intensity of FAD in the pure enzyme solution of sarcosine oxidase and the concentration of sarcosine oxidase protein, such as Figure 5 As shown, the regression equation I F =44.7+1674c, the R of the regression equation 2 = 0.9929.
[0057] 2. In order to verify the accuracy of...
Embodiment 3
[0062] Example 3. Using three-dimensional fluorescence spectroscopy to detect the presence of cholesterol oxidase in the crude enzyme solution of cholesterol oxidase
[0063] 1. Determination of the position of the endogenous fluorescence characteristic peak of the pure enzyme of cholesterol oxidase
[0064] The cholesterol oxidase gene (GenBank: MK757498.1) derived from Burkholderia cepacia ZWS15 was cloned into the vector pET28a, and the recombinant expression plasmid was transformed into E.coli BL21(DE3) for inducible expression. The constructed engineered bacteria E.coli BL21(DE3)-pET20b(+)-COD was single-colonized into the medium, cultured overnight at 37°C, 200rpm, inserted into LB medium, and cultured overnight at 37°C. Insert TB culture medium according to 5% inoculum amount, cultivate at 37°C, 200r / min until the cell concentration OD 600 After about 0.6-0.8, add 20% lactose induction solution, 28°C, 200rpm, induce culture for 20h. 8000r / min, centrifuge at 4°C for 5m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com