Bst DNA polymerase recombinant mutant, coding DNA thereof and ultrafast magnetic bead LAMP detection method
A detection method, polymerase technology, applied in DNA/RNA fragments, recombinant DNA technology, chemical instruments and methods, etc., can solve the problems of poor tolerance of high salt and inhibitors, poor extension ability, low sensitivity, etc., and achieve reversal The effect of improved transcriber activity, improved extension ability, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
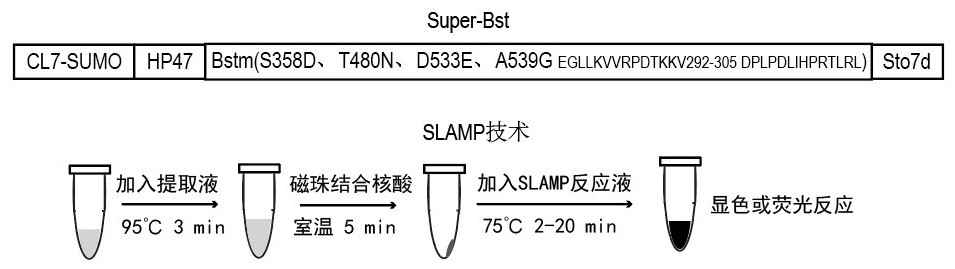
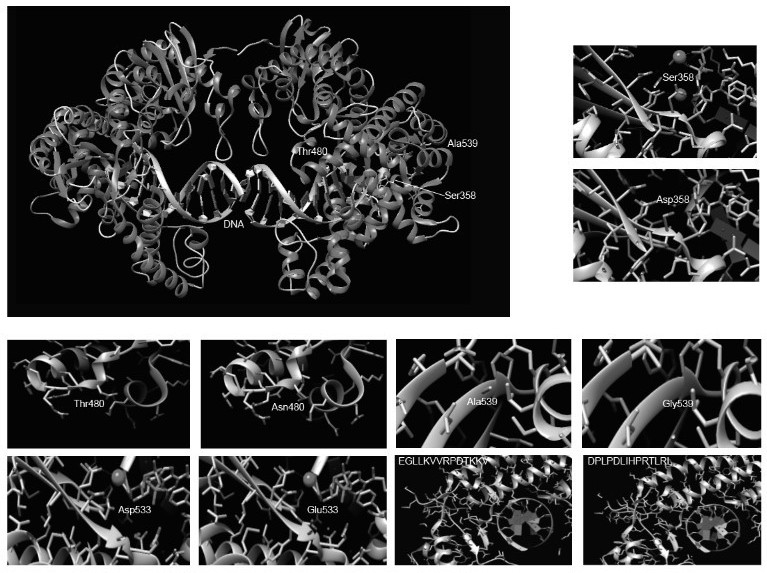
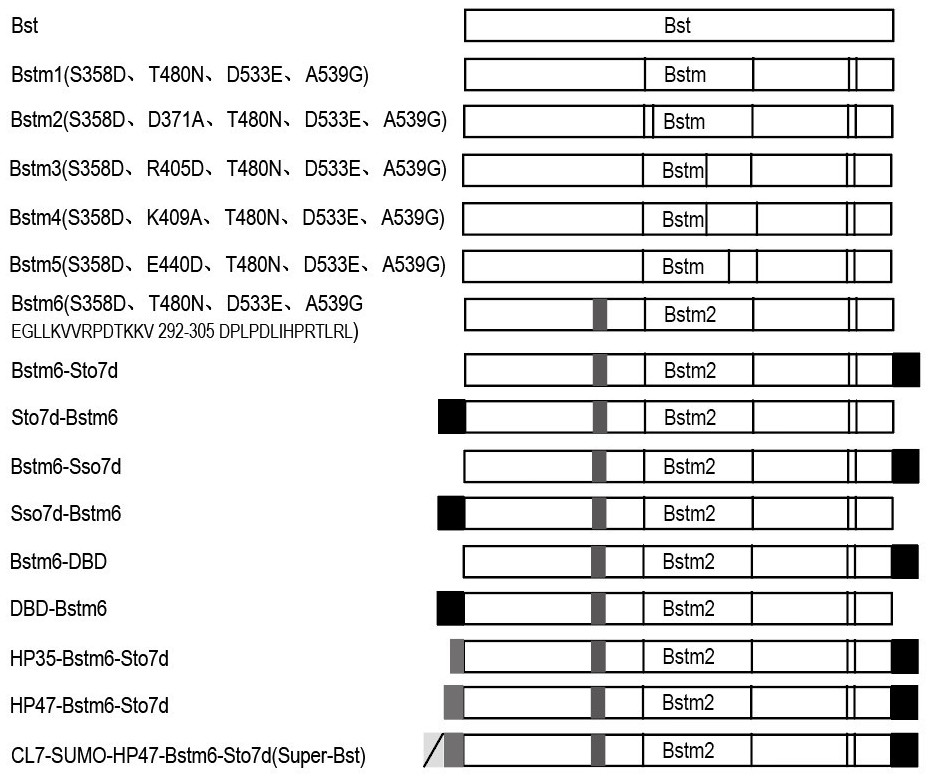
[0058] In this example, we selected some conserved active amino acid sites for mutation based on the three-dimensional crystal structure of Bst DNA polymerase and template DNA (PBD: 4SDJ) (such as figure 2 ), and fused with some functional domains that increase enzyme stability and elongation ability, see recombinant mutant protein image 3 , image 3 The 16 protein structures in the sequence correspond to SEQ ID No.1-16 from top to bottom. Subsequently, we tested the performance of the Bst mutant crude enzyme. The specific implementation is as follows:
[0059] 1) Preparation of Bst mutant prokaryotic expression strain
[0060] The synthesized Bst mutant DNA was connected to the pET-28a vector, and transformed into DH5α Escherichia coli competent, spread on the LB solid plate containing kanamycin, and cultured upside down at 37°C overnight.
[0061] Pick a monoclonal colony in good condition and culture it in LB liquid medium containing kanamycin at 37°C with shaking at ...
Embodiment 2
[0086] Example 2: Purification of Super-Bst.
[0087] In this example, we publish a specific protocol for the purification of Super-Bst.
[0088] 1) Super-Bst induced expression
[0089] According to the inoculation density of 1:200, BL21(DE3) Escherichia coli containing pET-28a-Bst mutant expression plasmid was inoculated into new LB medium containing kanamycin, and cultured with shaking at 37°C for 2.5 h until the OD600 value reached After 0.3, the shaking culture was continued at 18°C for 0.5 h until the OD600 value reached 0.3. Add IPTG at a final concentration of 0.3 mM, and culture with shaking at 18°C for 18 h. The cells were collected by centrifugation at 4000×g at 4°C. After washing the cells with pre-cooled PBS three times, weigh 3 g cells and add 30 ml of pre-cooled lysis buffer (20 mM Tris-HCl, 500 mM NaCl, 5% Glycerol, 0.1 mM PMSF, 1% protease inhibitors, pH 8.0) suspended bacteria. Process using a high pressure stirrer (~15,000 PSI) at 4°C for 3 min. Af...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com