Radish genome SNP-Panel and application thereof
A genome and radish technology, applied in the fields of application, plant genetic improvement, recombinant DNA technology, etc., can solve the problems of inability to large-scale application, inability to meet individualization, and high price of gene chips, to promote agricultural development, improve breeding efficiency, shorten the Effects of Breeding Cycles
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Extraction of Radish Genomic DNA
[0031] 1. Select radish tissue, preferably young tissue, including dry seeds, fresh leaves, young ears and old leaves, seedlings or young stems;
[0032] 2. Reagent preparation: CTAB buffer, including 2% CTAB, 1.4M NaCl, 100mM Tris-HCl, 10mM EDTA, pH=8.0; Washing buffer, including 76% ethanol, 10mM ammonium acetate; TE buffer, including 20mM Tris -HCl, lmMEDTA, pH8.0; ice absolute ethanol, pre-store absolute ethanol at -20°C for more than 1 hour.
[0033] 3. Use the CTAB method to extract DNA from radish tissue: cut the tissue from radish into pieces, and grind it in a liquid nitrogen environment; add preheated CTAB equivalent to the amount of the tissue, and quickly place it in a water bath at 65°C for 30 minutes Shake once every 5 minutes (preferably in a water bath for 1 hour); centrifuge at 4°C and 12000rpm / min for 10min, remove the supernatant and add an equal volume of chloroform and isoamyl alcohol (the volume ratio o...
Embodiment 2
[0034] Example 2 Design of amplification primers for specific SNP markers
[0035] 1. The selection of SNP markers, analyze the whole genome SNP (the version number of the whole genome sequence is R.sativus_genome_V1.1, the website is http: / / brassicadb.org / brad / datasets / pub / Genomes / Raphanus_sativus / ), by pressing Selecting one SNP variation per 1Mb distance to obtain a large number of SNP markers on the whole genome, wherein the number of a large number of SNP markers evenly distributed on the radish genome is 1000 or more;
[0036] 2. Amplification primer design, after sequence comparison, analysis and splicing, after converting the genome data into a database file, use Primer3 to design the amplification primers in batches, and use the NCBI-ePCR program to process the SNP-marked amplification primers one by one Test, screening can only amplify a single band containing the SNP marker, and locate the SNP marker at the chromosomal site where the template is located, and finally...
Embodiment 3
[0041] Example 3 Detection and Analysis of Individual Genotypes
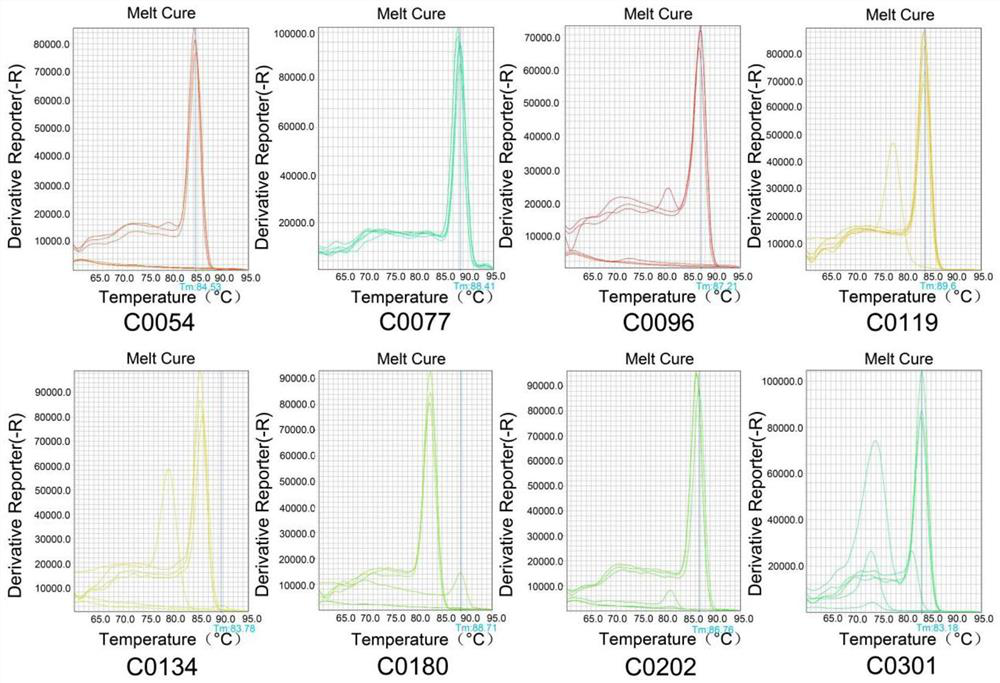
[0042] 1. Multiplex PCR amplification: the present invention relates to the amplification of 305 SNP markers. 305 SNP markers are combined to form a gene panel. Using the amplification primers of the 305 pairs of SNP markers, the extracted radish tissue DNA target The region is amplified; the amplified product is digested and an adapter is added, the fragment is selected after recovery and purification, and the construction of the next-generation sequencing library is finally completed;
[0043] 2. Quality control and quantification: including gel electrophoresis, real-time fluorescent quantitative PCR and Agilent 2100 for quality control and quantification of the constructed library, in preparation for on-machine sequencing;
[0044] 3. Complete PE150 sequencing on an illumina sequencer to obtain sequence data;
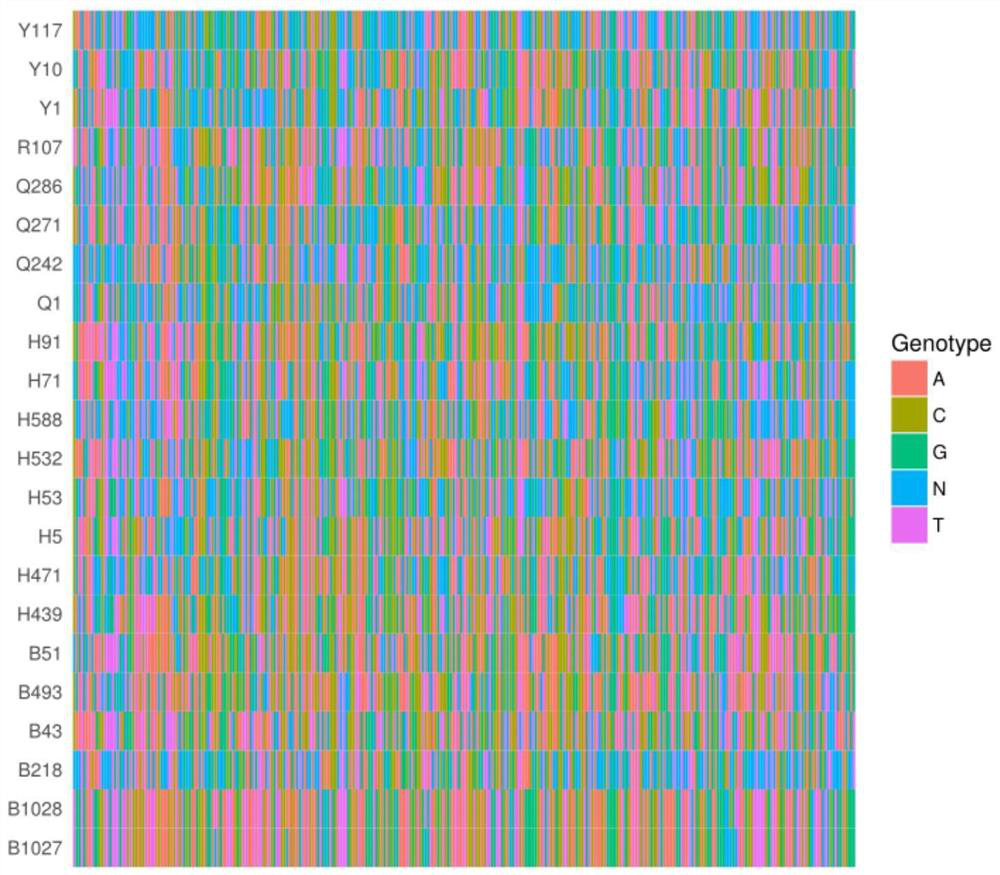
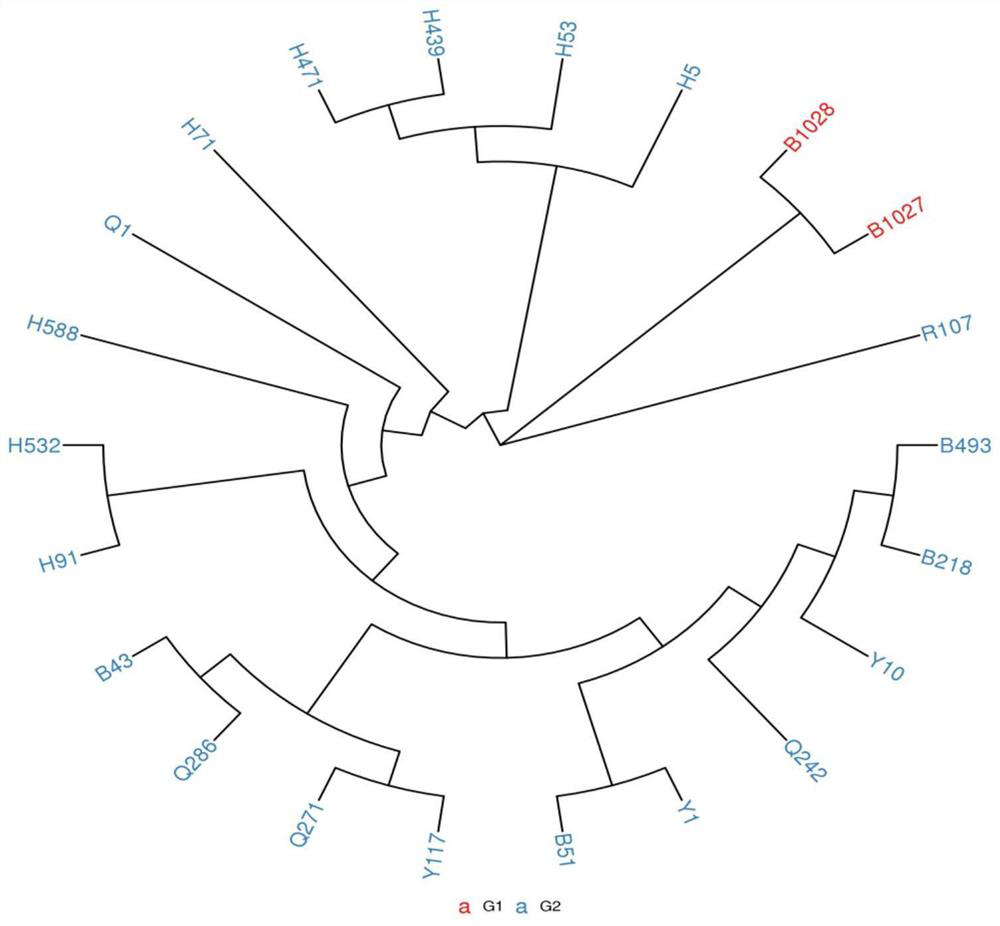
[0045] 4. Analyze the sequence data obtained by sequencing to obtain the SNP genotype result. Man...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com