Bacillus subtilis with inactivated extracellular protease as well as construction method and application of bacillus subtilis
A Bacillus subtilis and extracellular protease technology, applied in the field of genetic engineering, can solve the problems of unfavorable recombinant protein secretion and expression, loss of target protein yield, etc., and achieve good amino acid utilization ability, no nutritional deficiency, and improved transformation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Construction of CRISPR series plasmids
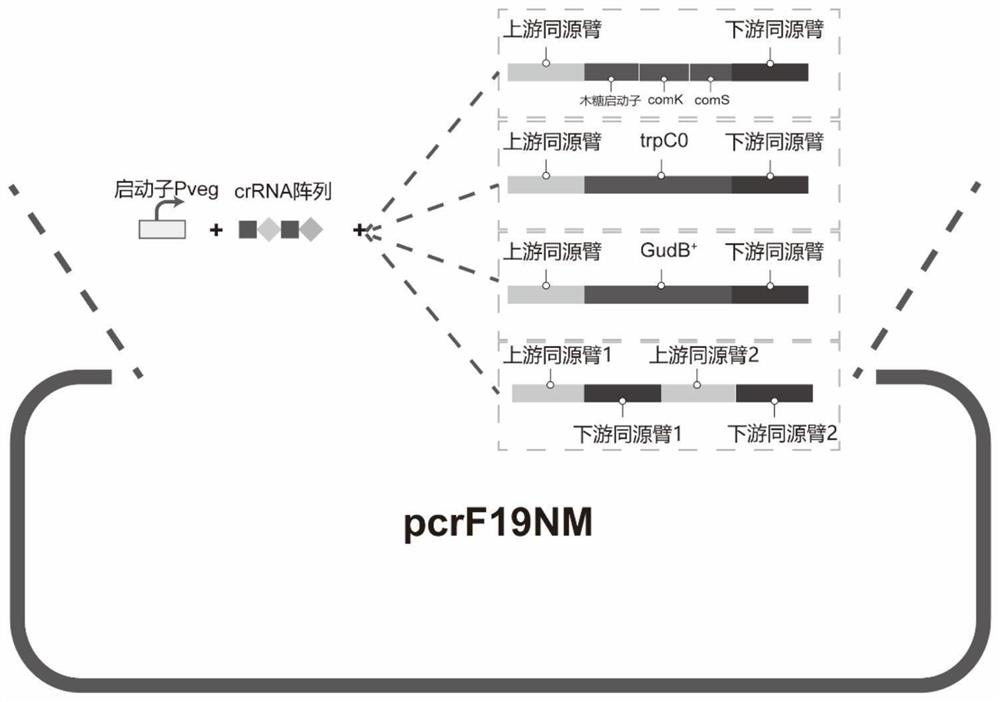
[0049] As described by Wu et al. in CAMERS-B: CRISPR / Cpf1 assisted multiple-genes editing and regulation system for Bacillus subtilis. Biotechnology and Bioengineering2020, 117:1817–1825, the CRISPR / Cpf1 system consists of two plasmids pHT-XCR6 and pcrF19NM2 (Available through the molecularcloud plasmid sharing platform, numbers MC_0068418 and MC_0101256, respectively). Plasmid pHT-XCR6 (ampicillin resistance in Escherichia coli, chloramphenicol resistance in Bacillus subtilis) is a Cpf1 expression vector, in which Cpf1 is induced and regulated by xylose; in addition, the plasmid also contains NgAgo protein Genes used to improve the efficiency of homologous recombination during gene editing. Plasmid pcrF19NM2 (both kanamycin resistant in Escherichia coli and Bacillus subtilis), and a temperature-sensitive plasmid in Bacillus subtilis, cannot replicate when cultured above 37°C, and is used as a crRNA expression vector ...
Embodiment 2
[0078] Embodiment 2: Construction of protease inactivated bacterial strain B.subtilis G601
[0079] The Cpf1 protein expression plasmid pHT-XCR6 of the CRISPR / Cpf1 system was transformed into a competent Bacillus subtilis, and spread on an LB plate containing chloramphenicol resistance until a single colony grew out. Then the bacterial strain transformed with pHT-XCR6 was made into a competent state, and transformed into the pcrF19-epr-xcomKS constructed in Example 1. After the plasmid was added to the competent state and cultivated for two hours, the plate was not directly coated, but the bacterial liquid Centrifuge (4000rmp, 2min) and resuspend in 500μL LB containing chloramphenicol, kanamycin and 3% xylose for overnight culture, then centrifuge and concentrate to 150μL the next day and add chloramphenicol and kanamycin LB plates with plain and 3% xylose. After a single colony grows, colony PCR can be performed to verify whether the gene editing has been completed. The suc...
Embodiment 3
[0080] Example 3: Verification of protease inactivated strain B.subtilis G601
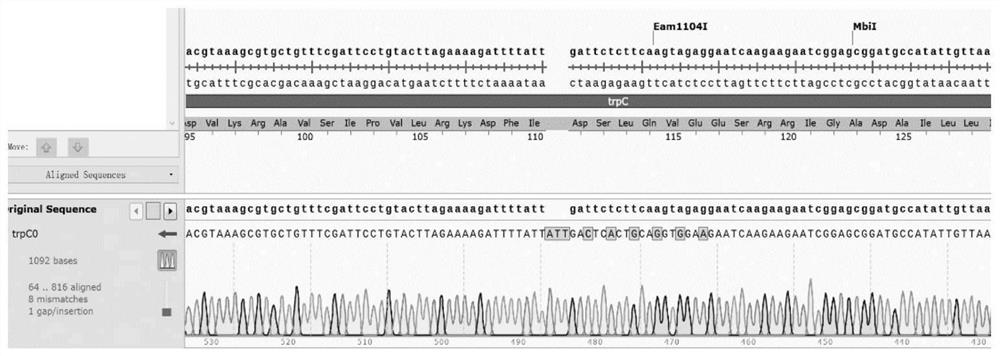
[0081] Such as image 3 As shown, the trpC0 on B. subtilis G601 was amplified and sequenced, which was consistent with the sequence of the designed mutation. Such as Figure 4 As shown, the wild-type strain B.subtilis 168 before mutation ( Figure 4 Upper) cannot grow on synthetic media, the mutated B.subtilis G601 ( Figure 4 Below) can grow normally. Such as Figure 5 As shown, put the gudB on the B.subtilis G601 + Amplified sequencing, which is consistent with the sequence of the designed mutation. Such as Figure 6 As shown, the engineered strains were subjected to colony PCR using gene name-VF and gene name-VR, and the results showed that B. subtilis G601 had integrated the xylose-inducible promoter P xylA The controlled transcription factor comK-comS gene was back-mutated to trpC0, and six extracellular protease genes were knocked out. Such as Figure 7 As shown, the ability of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com