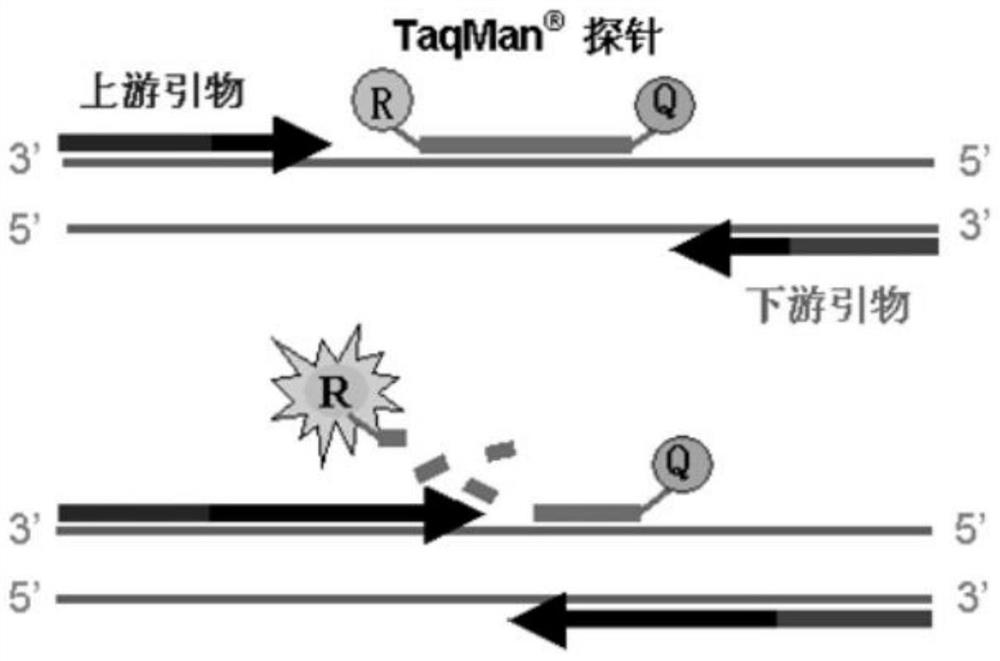
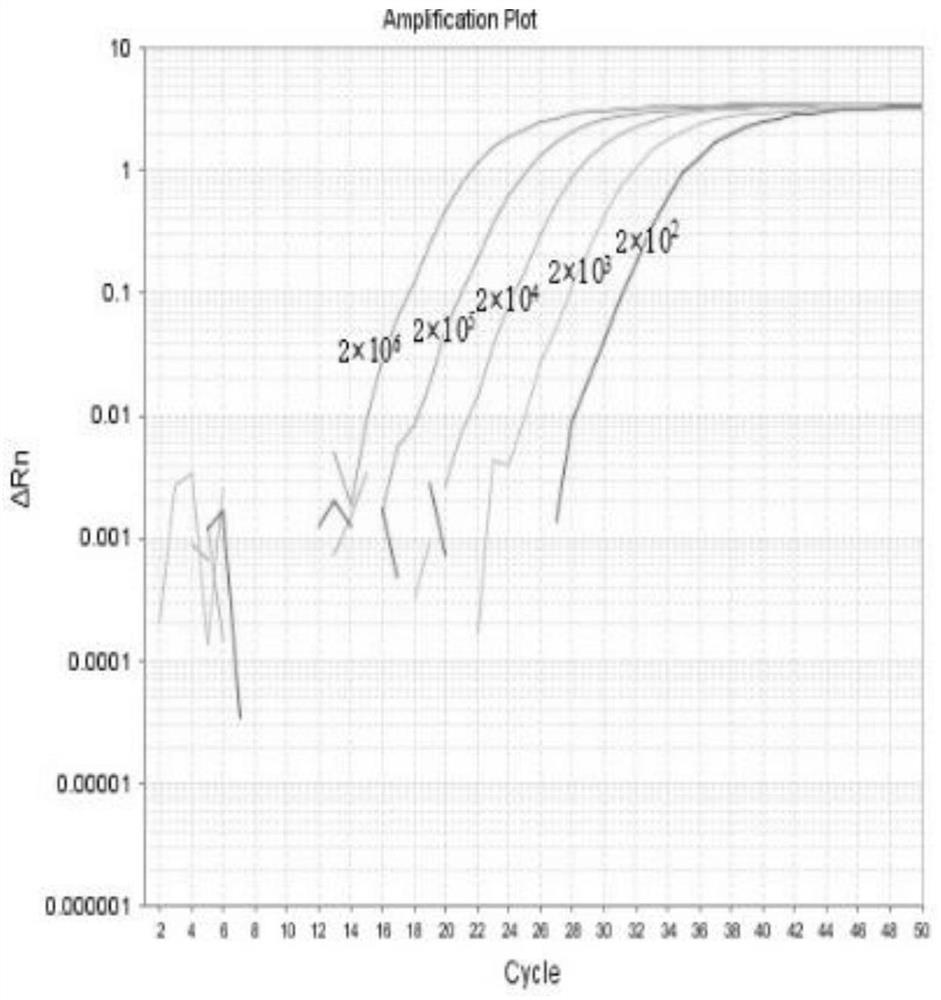
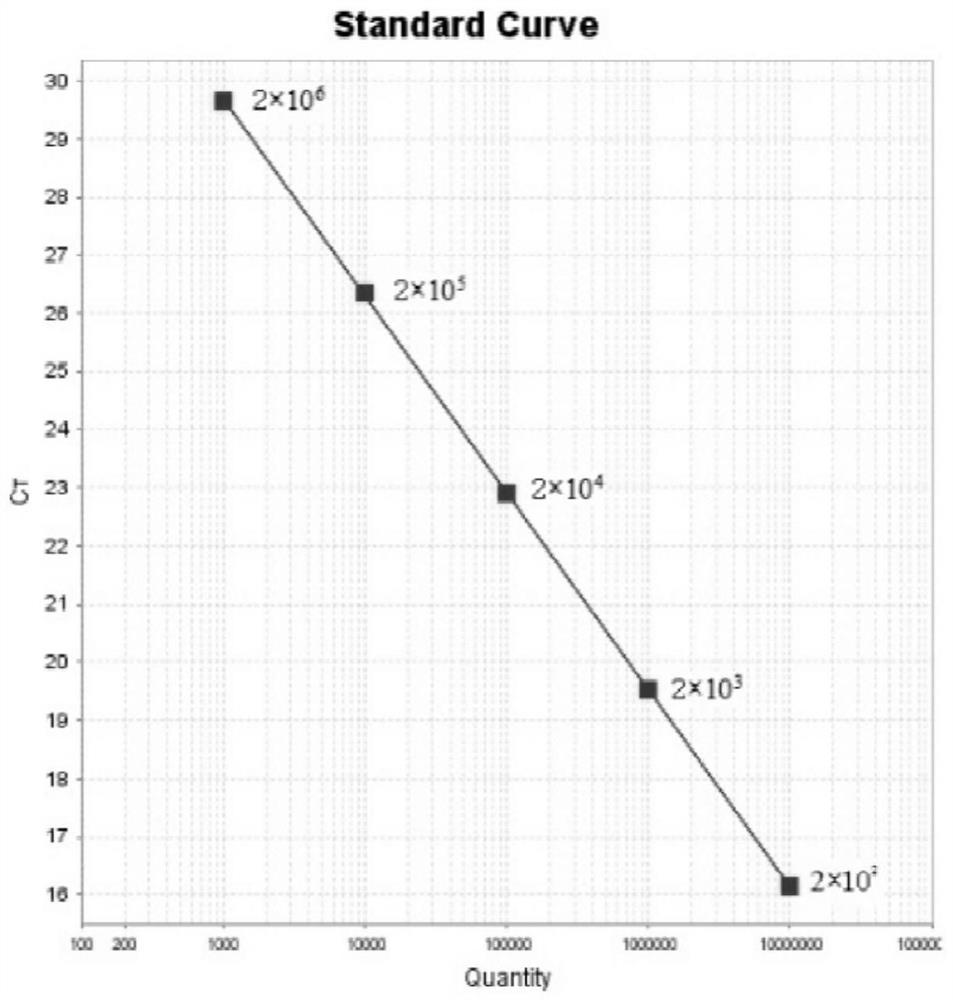
Fluorescent quantitative PCR detection method and kit for porcine circovirus type II
A porcine circovirus and detection kit technology, which is applied in the directions of microorganism-based methods, microbial determination/inspection, biochemical equipment and methods, etc. Good repeatability and stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 Preparation of PCV2 Amplified Product Recombinant Plasmid and Standard Item
[0050] Utilize NCBI database to find out the ORF2 gene of PCV2, take PCV2 (BJ0804 strain) ORF2 gene as the target sequence, GenBank accession number is EU921257.1, full-length 149bp, specific sequence is as shown in SEQ ID NO.1, carry out design primer and probe The needles were designed using the software Primer Premier 5.0 for primers and probes, and Oligo DNA man 4.0 and PrimerAnalysis were used for comparison analysis. The specific sequences are as follows:
[0051] PCV2-P1: 5'-CGGATATTGTATTCCTGGTCGTA-3';
[0052] PCV2-P2: 5'-CCTGTCCTAGATTCCACTATTGATT-3';
[0053] PCV2-Probe: FAM-5'-CTAGGCCTACGTGGTCTACATTTC-3'-TAMRA.
[0054] Viral genomic DNA of PCV2 was extracted using the viral genomic DNA / RNA extraction kit produced by Tiangen Biochemical Technology (Beijing) Co., Ltd.
[0055] Porcine circovirus type 2 strain BJ0804 was used as a standard template, primers P1 and P2 were ...
Embodiment 2
[0060] Embodiment 2 A kind of fluorescence quantitative real-time detection method of porcine circovirus type 2 (PCV2)
[0061] 1. Design primers
[0062] Use the NCBI database to find out the ORF2 gene of PCV2. Based on the PCV2 (BJ0804 strain) ORF2 gene as the target sequence, the GenBank accession number is EU921257.1, the full length is 149bp, and the specific sequence is shown in SEQ ID NO.1. Needle, using the software Primer Premier 5.0 to design primers and probes, Oligo DNA man 4.0 and Primer Analysis for comparison analysis, the specific sequence is as follows:
[0063] PCV2-P1: 5'-CGGATATTGTATTCCTGGTCGTA-3';
[0064] PCV2-P2: 5'-CCTGTCCTAGATTCCACTATTGATT-3';
[0065] PCV2-Probe: FAM-5'-CTAGGCCTACGTGGTCTACATTTC-3'-TAMRA.
[0066] 2. Preparation of reaction system
[0067] In order to obtain an efficient and sensitive quantitative PCR reaction system, different primers, probe (or dye) concentrations, Mg 2+ Concentration or annealing temperature is optimized, and t...
Embodiment 3
[0080] Fluorescence quantitative PCR detection method specific verification of embodiment 3PCV2
[0081] According to the method of Example 2, the cDNA of PCV2 plasmid, PCV2 virus sample, classical swine fever virus (CSFV), porcine respiratory and reproductive disorder syndrome virus (PRRSV), porcine pseudorabies virus (PRV) is respectively used as template and double distilled water to carry out Fluorescent quantitative PCR detection was used to determine the specificity of the detection method.
[0082] Test results such as Figure 4 As shown, the abscissa is the cycle number, and the ordinate is the fluorescence value, 1-3: 3 repeated experiments of PCV2 plasmid, 4-6: 3 repeated experiments of PCV2 virus samples, 7: 3 repeated experiments of CSFV virus samples, 8: 3 repeated experiments for PRRSV virus samples, 3 repeated experiments for 9: PRV virus samples, 3 repeated experiments for 10: negative control.
[0083] Depend on Figure 4 It can be seen that the primers P1,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com