Microfluidic detection system for identifying nontuberculous mycobacterium strains
A mycobacterium and non-tuberculosis technology, applied in the field of microbial detection, can solve problems such as unfavorable clinical and timely diagnosis, and achieve good clinical application prospects, high sensitivity, and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] The present embodiment provides specific detection of Mycobacterium intracellulare, Mycobacterium avium, Mycobacterium chelonae, Mycobacterium sulja, Mycobacterium simiensis, Mycobacterium marinum, Mycobacterium terrae, Gordon's species For the primer sets of Mycobacterium, Mycobacterium kansasii, and Mycobacterium fortuitum, a sequencing company is entrusted to synthesize the primers. The sequences of each primer set are:
[0054] (1) Mycobacterium intracellulare-specific detection primer set includes:
[0055] Forward outer primer F3: GGATGCGGGTCAAGTACG (SEQ ID NO: 1);
[0056] Reverse outer primer B3: CGTTATGCAGCGTGTTGAAG (SEQ ID NO: 2);
[0057] Forward internal primer FIP:
[0058] GTTTGAAGGCCACCTTGCGGTCTCCACCGAAGAATTCACCA (SEQ ID NO: 3);
[0059] Reverse inner primer BIP:
[0060] GCAGCTATCGCGACGTCGACCTCTCTCCTGGATACCTTCCT (SEQ ID NO: 4);
[0061] Forward loop primer LF: GCAGCGAGTTGATGAAGTCGT (SEQ ID NO: 5);
[0062] Reverse loop primer LB: GTGCTGCTCGTCGATGAC...
Embodiment 2
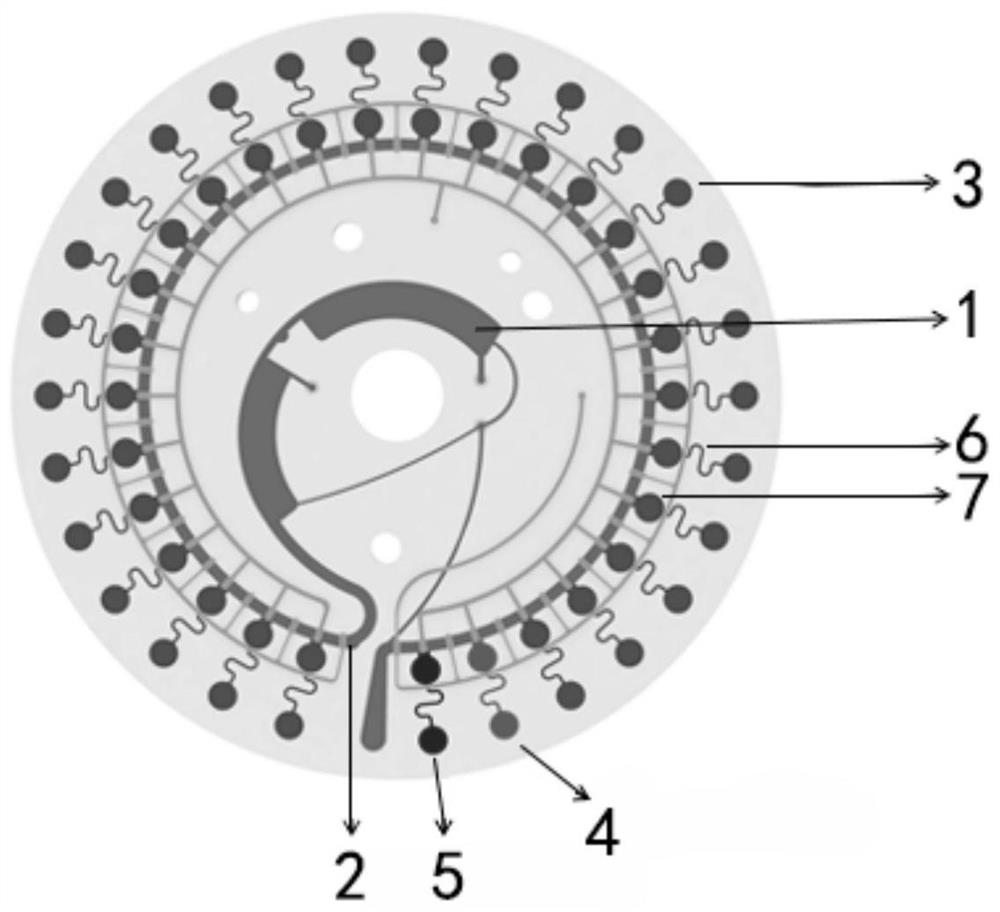
[0145] refer to figure 1 , this embodiment provides a microfluidic chip.
[0146] The microfluidic chip includes a chip body, and the chip body is provided with a sampling hole 1, a main channel 2, a microfluidic channel 6 and a reaction hole; the reaction hole includes a positive quality control hole 4, a positive control hole 5 and at least A detection hole 3; the sample loading hole 1 communicates with the main channel 2; each reaction well communicates with the main channel 2 through the micro flow channel 6; each of the detection holes 3 contains a specific non-tuberculous mycobacterium Sex detection primer set.
[0147] As a preferred solution, a buffer hole 7 is also provided between the reaction hole and the main channel 2 . By setting the buffer hole, so that the sample solution to be tested is filled with the buffer hole 7 first during the sampling process, and then enters the detection hole 3 or the positive quality control hole 4, and the positive control hole 5,...
Embodiment 3
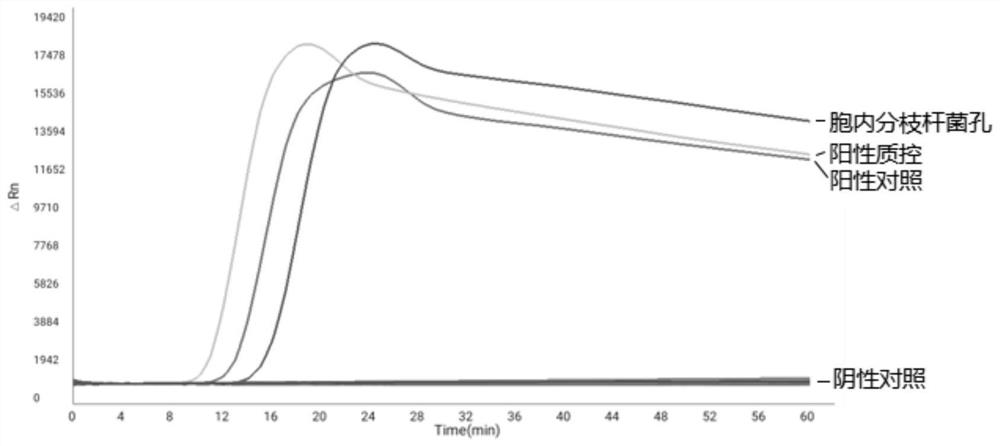
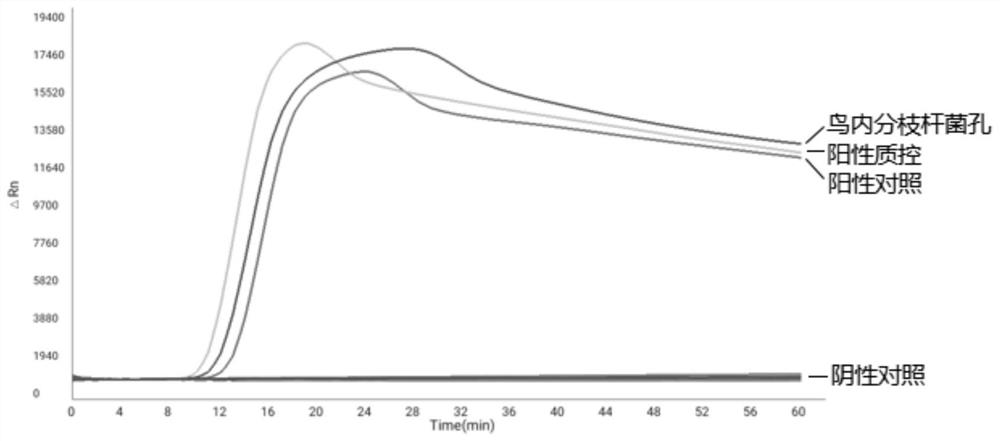
[0153] This embodiment provides a method for using the microfluidic chip provided by the present invention to perform specific detection of non-tuberculous mycobacteria, including:
[0154] (1) Genome extraction kits were used to extract Mycobacterium intracellulare, Mycobacterium avium, Mycobacterium chelonis, Mycobacterium sulja, Mycobacterium ape, Mycobacterium marinum, Mycobacterium terrae, and Gordon Genomic DNA of Mycobacterium, Mycobacterium kansasii, and Mycobacterium fortuitum was used as the template DNA to be tested.
[0155] (2) Prepare the reaction solution to be tested, including: 1 μL Tris (20mM), 1 μL KCL (10mM), 1 μL MgSO 4 (5mM), 1 μL NaH 2 PO 4 (10mM), 1μL Tween (0.5mM), 3.5μL dNTPs (1.5mM), 4μL betaine (1M), 1μL Bst enzyme, 0.5μL SYTO-9, 2μL test template DNA, 8μL sterilized water.
[0156] (3) Add the reaction solution to be tested prepared in step (2) to the sampling hole 1 on the microfluidic chip, so that the sample solution to be tested is distribut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com