Interaction identification and application of bovine skeletal muscle differentiation related DNA methylase TET1 and m6A methylase METTL3
A TET1-WT, methylation technology, applied in the field of molecular biology, can solve the problem that the relationship between epigenetic modifications is rarely found.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1: TET1 mRNA m during differentiation of bovine skeletal muscle myoblasts 6 A difference in methylation levels
[0070] The test materials were Qinchuan cattle myoblasts before and after differentiation m 6 The data after A-seq sequencing and the remaining m 6 A mRNA samples after immunoprecipitation.
[0071] The specific operation is as follows:
[0072] 1.m 6 Analysis and visualization of A-seq sequencing data
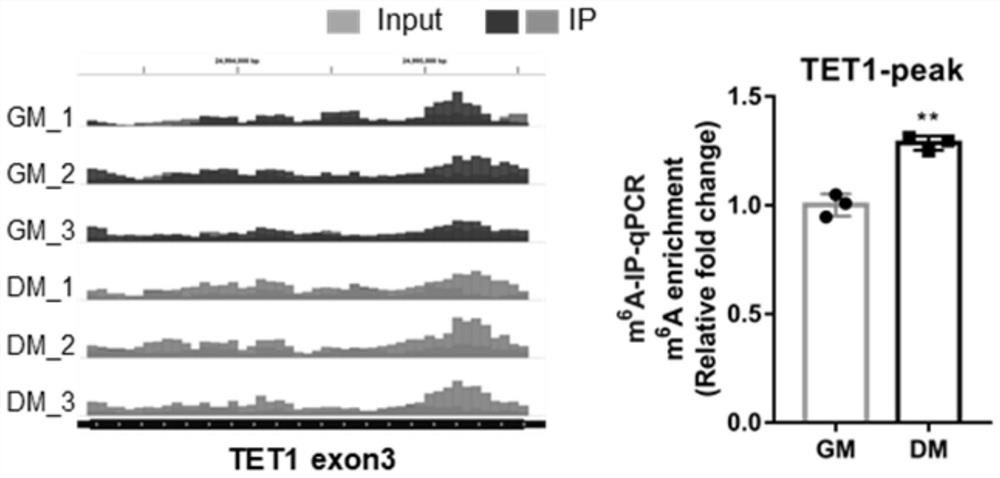
[0073] (1) Further bioinformatics analysis was performed on the sequencing data, and the m 6 A peak map visualization, indicating the m of TET1 mRNA before and after myogenic differentiation 6 A methylation position and m 6 There are differences in the methylation level of A ( figure 1 left picture);
[0074] (2) According to the m of TET1 in the sequencing results 6 A peak sequence, at m 6 A modification site prediction website (http: / / www.cuilab.cn / sramp) to determine the m of TET1 mRNA 6 The A site is located at the 3999th nucleotide of...
Embodiment 2
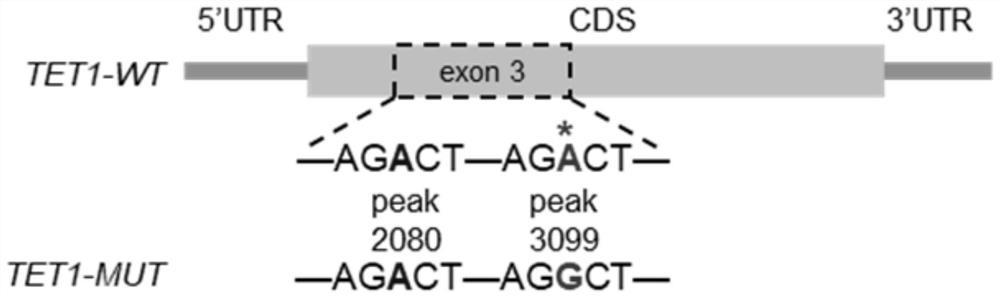
[0081] Example 2: Changing the m of TET1 mRNA 6 A method of methylation modification, that is, for m 6 A site for synonymous mutation;
[0082] The nucleotide sequence of the coding region of the bovine TET1 gene is as follows:
[0083] atgtctcgat ctcgccacgc aagaccttcc aaattagtca ggagggaaga tctaaacaag 60
[0084] aagaaaaaca accaactgaa gaagggatct aagccagcca acaaaaatgt ggcgactgtc 120
[0085] aagactgtga gccctggaaa attgaagcaa ttcgtgcaag aaagagatgt taagaaaaaa 180
[0086] acagaaccca aactgactgt gccagtcaga agccttctga caagagctgg agcagcacga 240
[0087] atgaatttgg ataggaccga ggttcttttt cagaacccag agtccttaac ttgcaatggg 300
[0088] tttacaatgg ctctgcgaag cacctctctt agcagacgac tctcccaacc cccaatggtc 360
[0089] atagccagac ccaagaaagt tccaccacct aagaatctag gaaagcaaca tgagtgtaat 420
[0090] tataaggtac ttacagacat gggagtaaag cactcagaaa atgattcagt cccaatgcaa 480
[0091] gaccccccag tcccccctga catagagaag ctaattggca tacaaaatgc atctttcatt 540
[0092] aaagacgaga gccaagagac aacccagtct tg...
Embodiment 3
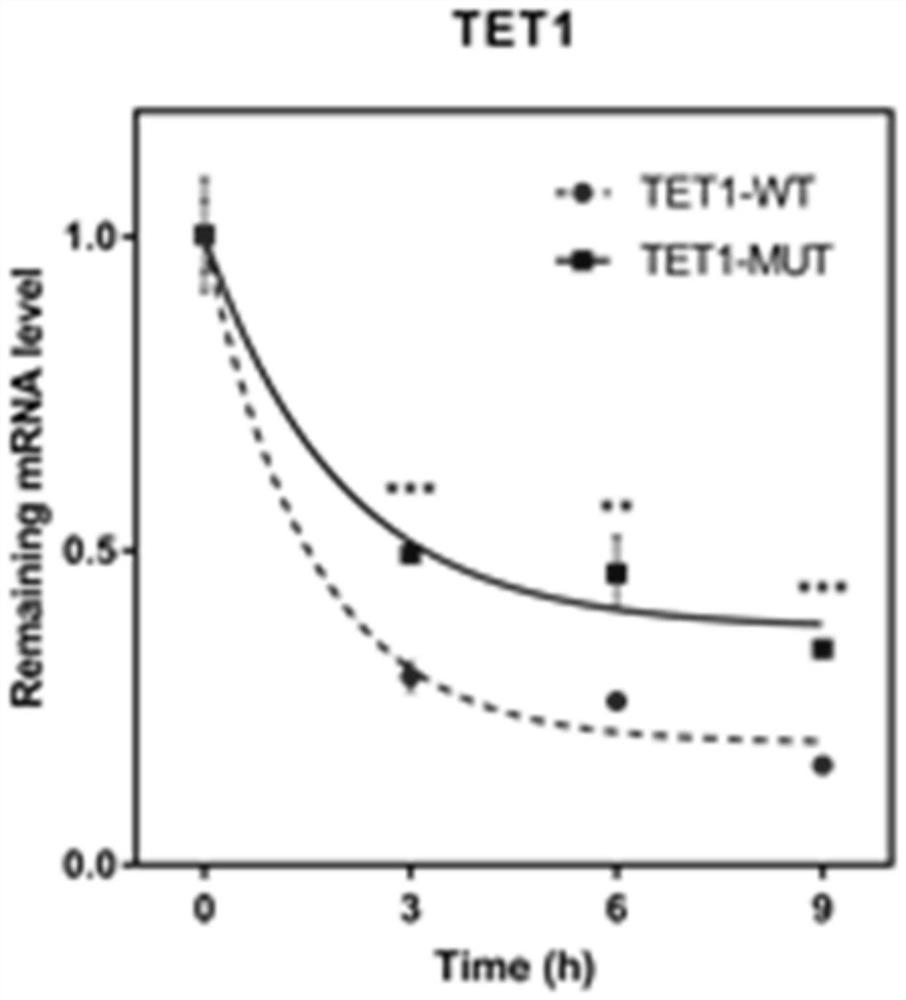
[0191] Example 3: TET1 mRNA m 6 The effect of A methylation site mutation on the stability of TET1 mRNA
[0192] (1) Cloning the coding region sequence of bovine TET1 gene ((ENSBTAT00000086206.1) and the sequence after synonymous point mutation into pcDNA3.1-3xFlag-EGFP-C mammalian expression vector to obtain wild type (MEF2C-WT) and mutant (MEF2C-MUT) expression plasmids;
[0193] (2) When the Qinchuan bovine skeletal muscle myoblasts grow to a density of 80-90%, subculture the cells into a 6-well plate;
[0194] (3) When the myoblast grows to 80% density, use the Lipofectamine 3000 kit (Invitrogen) to transfect the MEF2C-WT and MEF2C-MUT expression plasmids into the myoblast, mainly refer to the instructions of the Lipofectamine 3000 kit, the specific operation as follows:
[0195] 1) For each well of cells, add 5 μl of p3000 reagent and 2500ng of plasmid DNA to 120 μl of OPTI-MEMI medium;
[0196] 2) For each well of cells, add 5 μl Lipofectamine 3000 reagent to 120 μl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com