CRISPRCas12a system-based miRNA-21 detection method and kit
A miRNA-21 and detection method technology, applied in the field of biological detection, can solve problems such as time-consuming, complex data analysis, poor primer versatility, etc., and achieve the effects of improving detection sensitivity, simplifying detection process, and improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
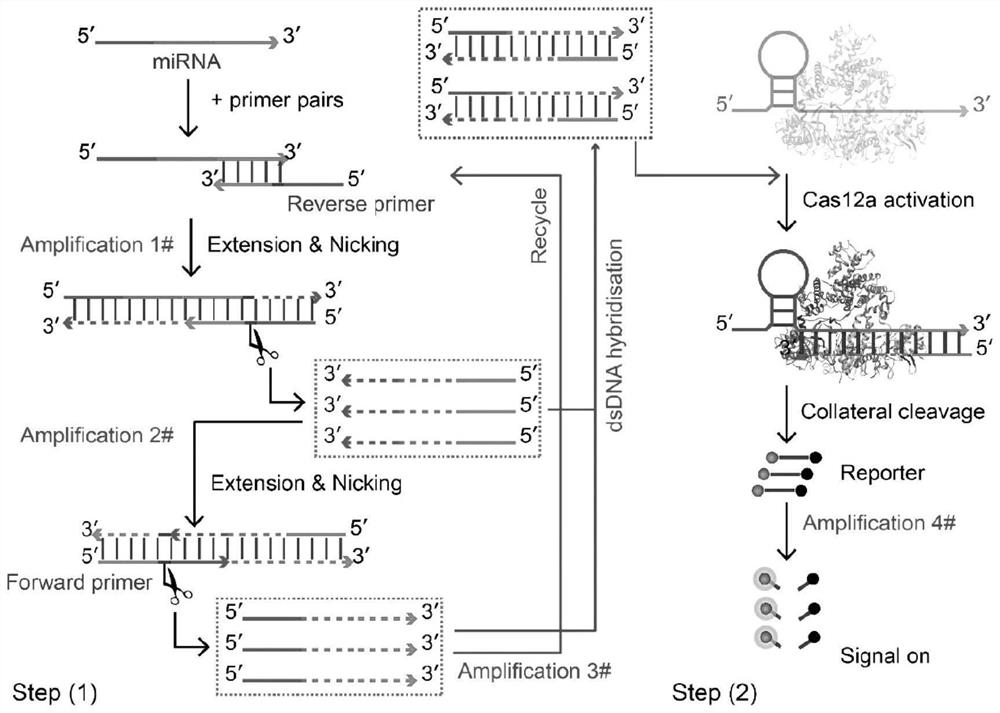
[0025] A miRNA-21 detection method based on CRISPRCas12a system, including the following steps:
[0026] Step 1: Synthesize a pair of specific incision primers according to the sequence of the miRNA-21 target to be measured, and edit the corresponding guide RNA sequence according to the CRISPR gene.
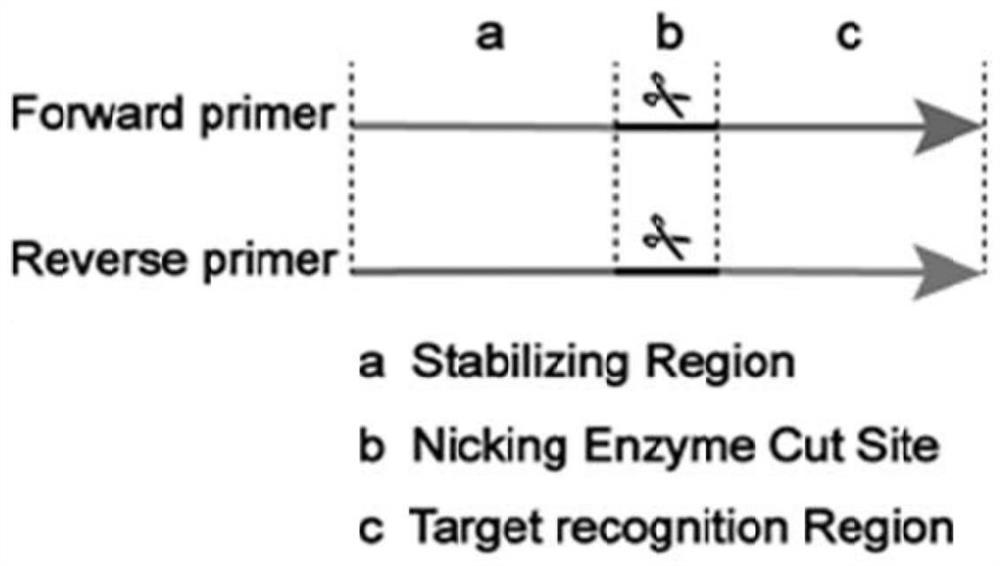
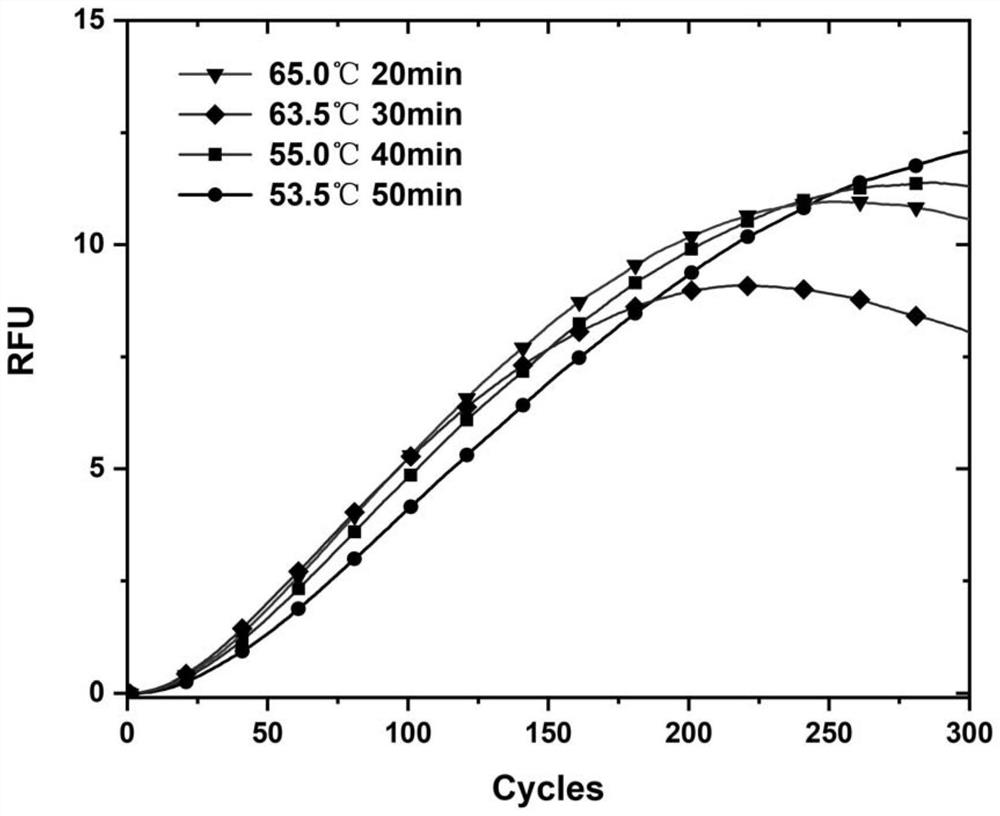
[0027] Designing the corresponding forward and reverse incision primers for miRNA targets requires that the primer design contain at least three parts, namely the strand displacement fixation region, the incision enzyme recognition region, and the target recognition region from the 5' to 3' ends, respectively, such as Figure 1 as shown. The stable combination of the primer fixation region and the complementary chain is the basis for the efficient implementation of double chain replacement, and it is also the key to improving the detection sensitivity, if the TM value of the fixed region is too low, it will lead to the primer detaching from the complementary chain, and the amplificati...
Embodiment 2
[0057]One kit comprises: primers synthesized according to the method of Example 1, guide RNA, nonspecific fluorescent reporter molecules, DNA / RNA polymerase, incisionase, Cas12a nuclease, dNTP, negative control DEPC-H 2 O as well as reaction buffer. The reaction buffer is a mixed solution of potassium acetate (25-75 mM), magnesium acetate (5-20 mM), bovine serum albumin (50-150 μg / ml), Tris-acetic acid (10-30 mM). The Ph of the mixed solution is 7.5-8.5.
[0058] Among them, the non-specific fluorescent reporter molecules are modified with quenching groups and fluorophores at both ends. The types of quenched groups are Dabcyl, BHQ-1, QYS-7, BHQ-2, and the fluorophores are Pacific Blue, Oregon Green, Bodipy FL-X, FAM, TET, Bodipy R6G-X, JOE, HEX, Cy3, Cy3.5, Cy5.5, Rhodamine Red-X, TAMRA, Texas Red-X, Any one of the ROX.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com