Chip for capturing, counting and lossless release of circulating tumor cells and application of chip
A tumor cell and chip technology, applied in the field of functional nanomaterials and biological detection, can solve the problems of insufficient strategies for sensing, detecting and counting circulating tumor cells, difficulty in non-destructive release of circulating tumor cells, and low abundance of circulating tumor cells, achieving excellent space Positioning ability, obvious affinity performance, effect of improving capture ability and effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1 Preparation of chips for capturing, counting and non-destructive release of circulating tumor cells
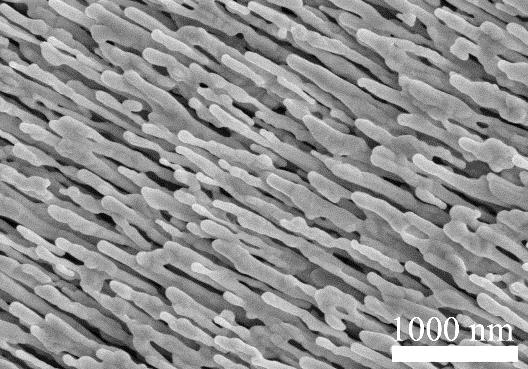
[0075] The first step is to use the oblique angle deposition method to prepare the evaporation coating technology ( [3] Liu Y J, Chu H Y, Zhao YP. Silver Nanorod Array Substrates Fabricated by Oblique Angle Deposition: Morphological, Optical, and SERS Characterizations [J]. The Journal ofPhysical Chemistry C, 2010, 114(18): 8176-8183). A 1 cm × 1 cm glass slide was placed in a vacuum deposition chamber, and 20 nm Ti, 200 nm Ag film, and 2000 nm film were sequentially deposited at the rates of 0.2 nm / s, 0.3 nm / s, and 0.3 nm / s, respectively. Ag film to form silver nanorods with morphologies such as figure 1 shown. The whole evaporation process is under pressure -6 Torr under high vacuum conditions.
[0076] In the second step, PDMS small holes with a diameter of 4 mm and a height of 1 mm were prepared and pasted on the silver nanorod array substrate to obtai...
Embodiment 2
[0083] Example 2 Characterization of the working mechanism of the chip for capturing, counting and non-destructive release of circulating tumor cells
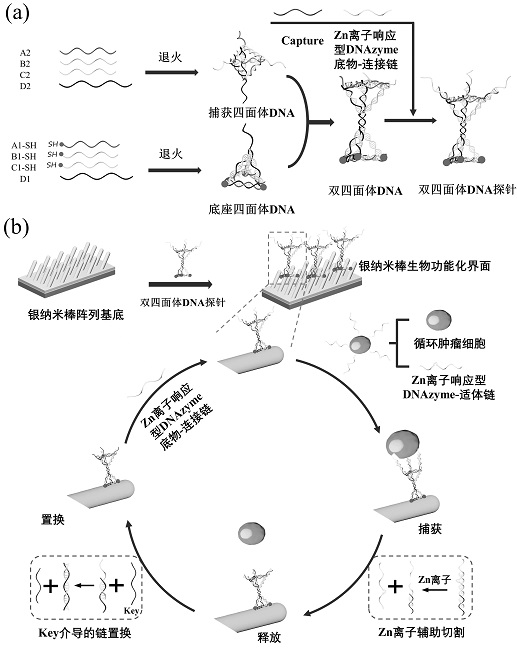
[0084] 1. To characterize the working mechanism of the chip prepared in Example 1 for capturing, counting, and nondestructively releasing circulating tumor cells. like figure 2 As shown in (b), the constructed double-tetrahedral DNA probe is covalently bound to the silver nanorod through three thiol groups on the bottom surface of the base tetrahedral DNA to form a biorecognition interface with nano-rough surface features. .
[0085] When capturing and releasing circulating tumor cells, first, a Zn ion-responsive DNAzyme-aptamer chain (which can specifically target and bind to cell surface membrane proteins) was co-coated with circulating tumor cells (human gastric cancer cell SGC-7901 as an analytical model). After incubation, the aptamer is modified to the surface of circulating tumor cells by specific binding of the aptam...
Embodiment 3
[0093] Example 3 Characterization of cell-on-chip capture performance for the capture, counting, and non-destructive release of circulating tumor cells
[0094] 1. In the case of ensuring that the Zn ion-responsive DNAzyme-aptamer chain is in excess relative to the number of cells, the Zn ion-responsive DNAzyme-aptamer chain was co-incubated with the target cell SGC-7901 to obtain Zn ion-responsive DNAzyme- Aptamer chain-labeled SGC-7901 cells.
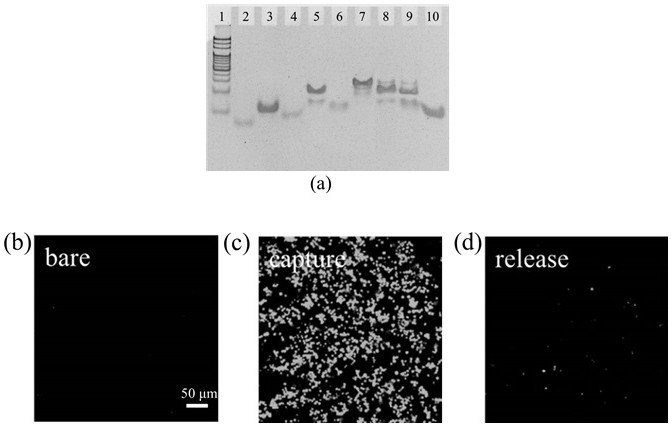
[0095]2. Co-incubate the chip with SGC-7901 cells bound with Zn ion-responsive DNAzyme-aptamer chains to capture and separate cells. In order to determine the optimal cell capture time, the cell capture performance of the nanobiointerface with different incubation times from 0 to 50 min was experimentally investigated. Figure 4 The middle (a) is the fluorescence imaging characterization of the captured cells for 0~50 min, Figure 4 The middle (b) is the corresponding capture efficiency statistics. With the increase of incubation ti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com