Flavone synthase I/flavanone-3-hydroxylase and application thereof in field of flavonoid compound synthesis
A flavanone, hydroxylase technology, applied in the directions of application, microorganism-based method, angiosperms/flowering plants, etc., can solve problems such as flavonoid synthase I and flavanone-3-hydroxylase have not been reported yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
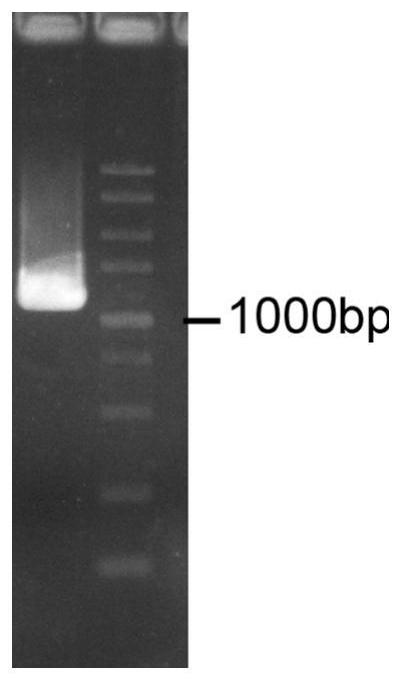
[0060] Example 1 Cloning of the expression gene PnFNS I / F3H
[0061] 1.1 CTAB-PVP method to extract total RNA of P.
[0062] The preparation method of CTAB-PVP extraction buffer is as follows:
[0063] 100 mM Tris·HCl (pH 8.0), 2% CTAB (w / v), 2% PVP (w / v), 25 mM EDTA, 2 M NaCl, mercaptoethanol was added to 0.2% after autoclaving; solution preparation was treated with DEPC ddH 2 O, spare after autoclaving.
[0064] Extraction Method:
[0065] (1) The water bath was adjusted to 65°C, and the prepared CTAB extract was placed in it to preheat. Take an appropriate amount of material into a mortar, add liquid nitrogen and grind into powder.
[0066] (2) Put the ground material into a liquid nitrogen quick-frozen imported 2 mL centrifuge tube, add 600 μL of CTAB extract, and invert up and down to mix evenly.
[0067] (3) 65 ℃ water bath, mix once every 10 minutes, and heat for 30 minutes.
[0068] (4) Take out the centrifuge tube and wait for the temperature to drop to room te...
Embodiment 2
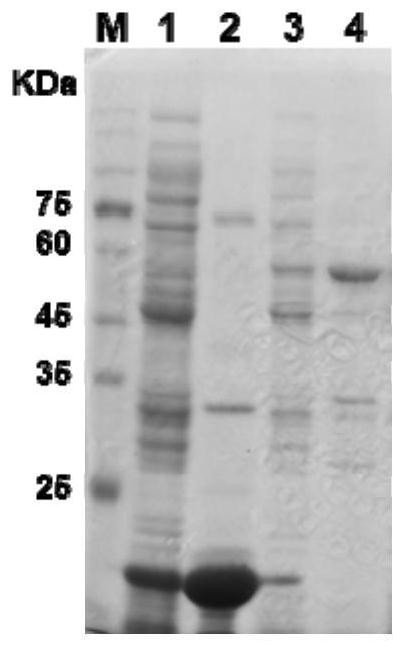
[0120] Example 2 Gene protein expression and enzyme activity function analysis
[0121] 2.1 Construction of protein expression vector
[0122] 2.1.1 Amplify the target gene
[0123] The correctly sequenced monoclonal bacteria were cultured overnight at 37°C at 120rpm, and the plasmid was extracted using the Plasmid Mini Kit:
[0124] (1) The existing strains MemUGT1-pTOPO-DH5α were divided into LB plates (containing 100 μg / mL Amp), 37 ° C, after 12 h, single clones were grown, and single clones were picked in 4 mL of Amp-resistant medium, 37 ° C , 110rpm cultured for 10h.
[0125] (2) Centrifuge the bacterial solution at 12,000 rpm for 1 min at room temperature, discard the supernatant, collect the bacterial cells, and discard the supernatant as much as possible.
[0126] (3) Add 150 μL of solution P1 to the centrifuge tube with the bacterial cell precipitation left, and vortex to shake until the bacterial cell is completely suspended.
[0127] (4) Add 150 μL of solution P...
Embodiment 3
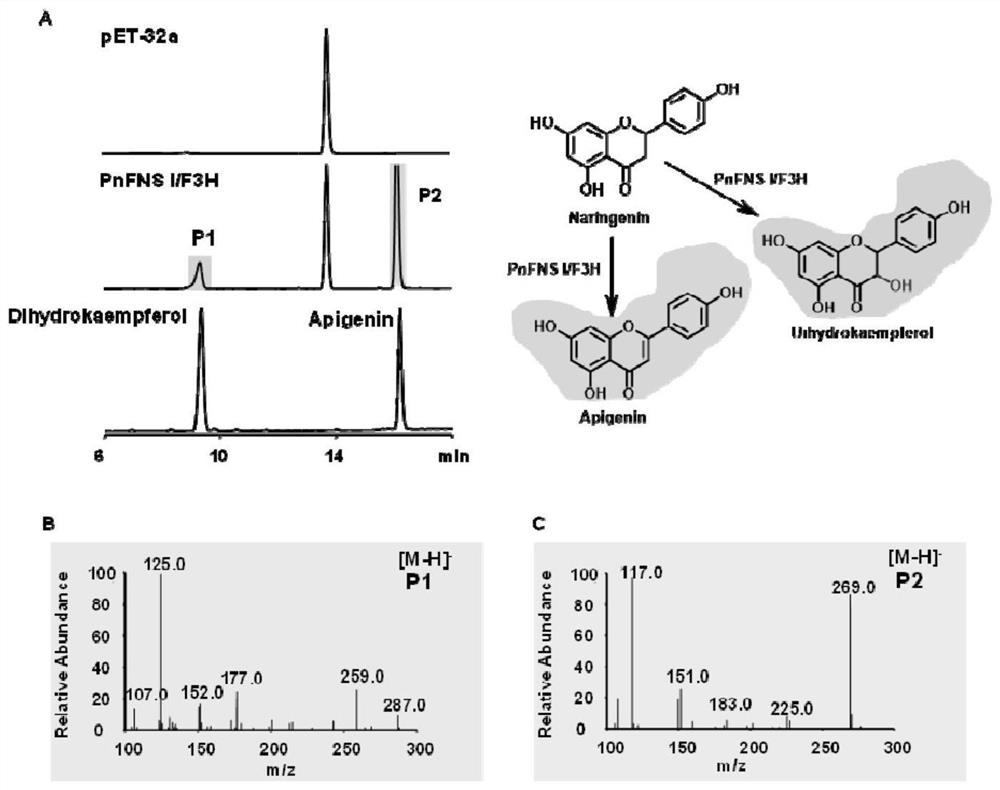
[0186] Example 3 Biosynthesis of apigenin and dihydrokaempferol using Escherichia coli PnFNS I / F3H-pET32a-BL21.
[0187] (1) Activate the strain in a constant temperature incubator at 37 °C, pick a single clone and inoculate it into 4 mL of LB liquid medium (containing 100 μg / mL of Amp), and continue to cultivate in a 37 °C incubator for 7 hours;
[0188] (2) The target strain and the control strain were inoculated into 50 mL of resistant LB medium according to the ratio of 1:100, and cultured at 37°C in a shaker at 200 rpm to OD600=0.6-0.8, and IPTG was added to make the final concentration 0.5 mM , 20 ℃ constant temperature incubation 6-7h;
[0189] (3) Add the DMSO-dissolved substrate (naringenin) to the bacterial solution, the substrate concentration is 150 μM, and put it into 20°C to continue culturing for a period of time;
[0190] (4) Take out 500μL of bacterial liquid every 12h, add an equal volume of ethyl acetate to extract 2-3 times, combine the organic phases, blo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com