Alfalfa Na+/H+ reverse transport protein gene and its clone and use
An antiporter, alfalfa technology, applied in the field of molecular biology and biology, can solve problems such as less obvious effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach 1
[0075] Embodiment 1: Alfalfa Na + / H + Cloning method of antiporter gene
[0076] (1) Extraction of RNA: Total RNA was extracted by CTAB method or RNA kit.
[0077] (2) Synthesis of the first strand of DNA: take 1 μg of total RNA, add 4 μl of 5× reaction buffer, 2 μl of 10 mM deoxyribonucleic acid (dNTP), 0.5 μl of ribonuclease inhibitor (40-200u / μl), primer oligodT (1 μg / μl) 1 μl, reverse transcriptase (10u / μl) 2 μl, react at 42°C for 60 minutes, then place at 85°C for 10 minutes to terminate the reaction.
[0078] (3) PCR reaction: polymerase chain reaction (PCR) reagents and conditions are:
[0079] First mix the following reagents together:
[0080] 10×Reaction Buffer 5μl
[0081] Deoxynucleotide mixture (dNTP) 4μl
[0082] Forward primer (5μM) 4μl
[0083] Reverse primer (5μM) 4μl
[0084] Template cDNA 4μl
[0085] Taq DNA polymerase 0.5μl
[0086] Total volume 50μl
[0087] The PCR reaction conditions are: 94°C for 3 minutes; then enter the following cycle: 9...
Embodiment approach 2
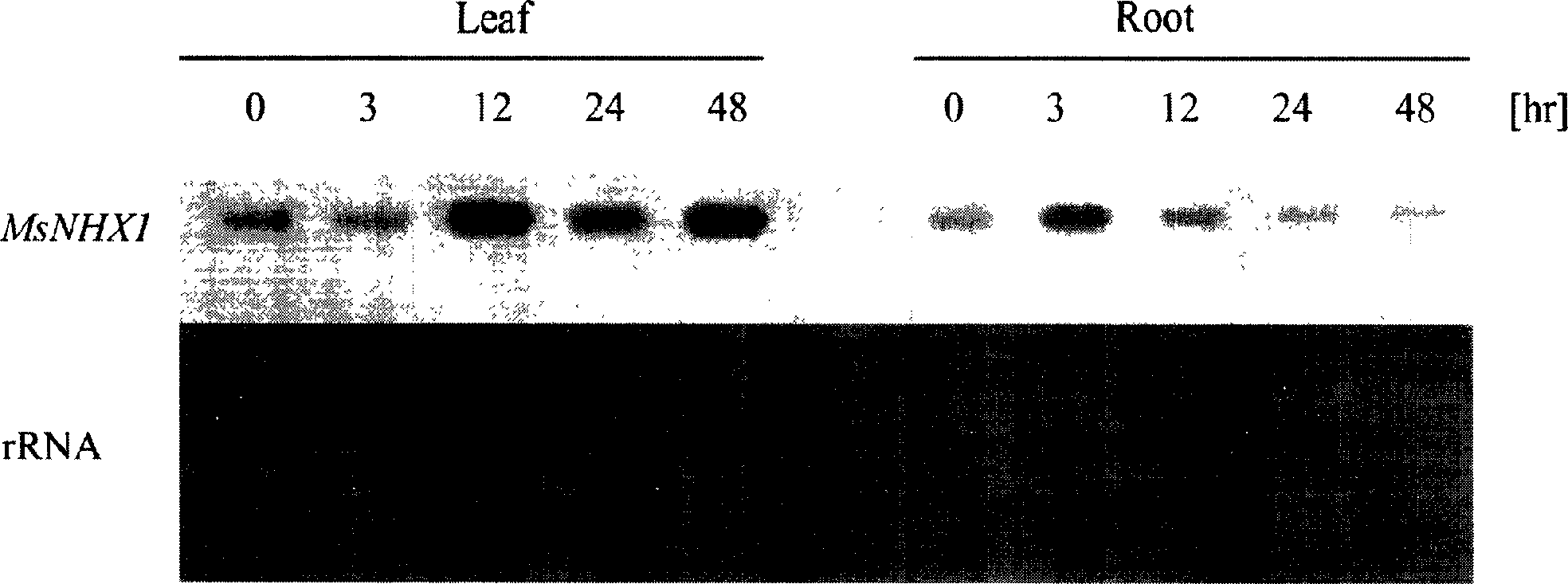
[0093] Embodiment 2: Alfalfa Na + / H + The sequence of the antiporter gene (MsNHX1) is shown in the "Summary of the Invention" section.
Embodiment approach 3
[0094] Embodiment 3: Construction of expression vector
[0095] (1) According to the separated Na + / H + Nucleotide sequence of antiporter gene, design primers:
[0096] Forward primer: 5′-TATTCTAGACGAGGTGGCGACCGGCATGG-3′
[0097] Reverse primer: 5′-GACGAGCTCCTTAACTACGGTCTTCTGC-3′
[0098] The polymerase chain reaction was performed using the cDNA reverse-transcribed from the root total RNA as a template.
[0099] (2) Take 2 μl of the PCR product and connect it to the pGEM-T easy vector, and the operation steps are carried out according to the instructions of pGEM-T easy and pGEM-T easy Vector system produced by Promega Company. Then Escherichia coli DH5α strain was transformed and grown overnight on LB plates coated with 5-bromo-4-chloro-3-indole-β-D-galactoside and X-gal containing ampicillin (100 μg / ml). Pick white colonies and culture them overnight in LB liquid medium. Then the plasmid DNA was extracted by alkaline method and sequenced.
[0100] (3) Cut the gene fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com