Plasmid with bar streptomycete Lat gene loss, derivative and constructing method thereof
A technique for gene deletion of Streptomyces clavicularis, applied in the field of bioengineering, can solve the problems of no single crossover integration into chromosome, lat gene deletion mutation, cumbersome steps, etc., and achieve the construction method is simple and easy, with strong purpose and high work intensity small effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
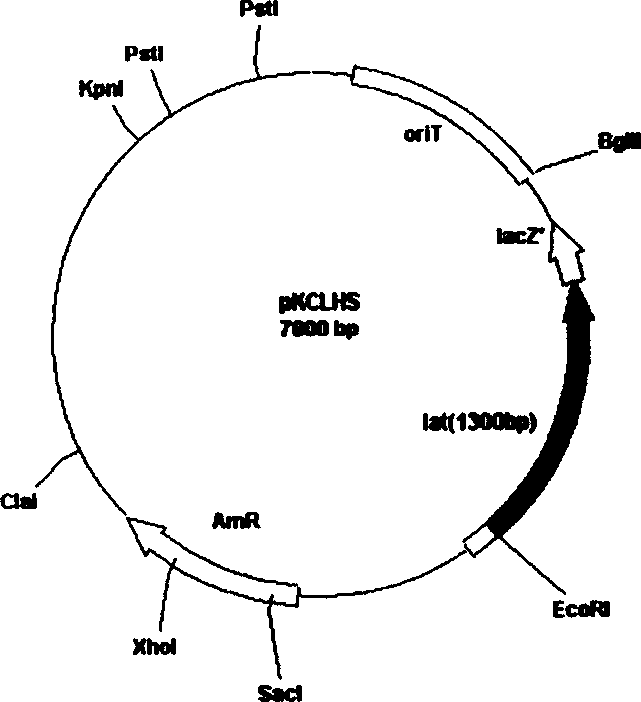
[0036] Construction of recombinant plasmid pKCLHS
[0037] ① Extraction of total DNA of Streptomyces clavulatus
[0038] Pick the solid ISP (ISP (Difco): 0.8%, agar powder: 2%, pH = 7.3) plate Streptomyces clavulatus cultured at 28°C for 3 days and inoculate it in the liquid ISP medium, culture it on a shaker at 28°C for 2 days, centrifuge The bacterial cells were harvested, and the bacterial cells were washed once with 10 mM EDTA at pH=8.0 or twice with water. Add SET (10mmol / L Tris-HCl (pH8.0), 10mmol / L NaCl, 1mmol / L EDTA (pH8.0)) solution to 1mL, and add 20μL of 50mg / mL (or an equivalent amount of other concentrations of lysozyme ), 37°C water bath for 30-60min. Add 28 μL of 20 mg / mL proteinase K and mix well, add 120 μL 10% SDS, invert and mix well, and incubate at 55° C. for 2 hours. Divide the mixture into two clean centrifuge tubes (0.8mL / tube), add 500μL of phenol / chloroform respectively, shake vigorously and mix thoroughly, and carefully draw the supernatant into a...
Embodiment 2
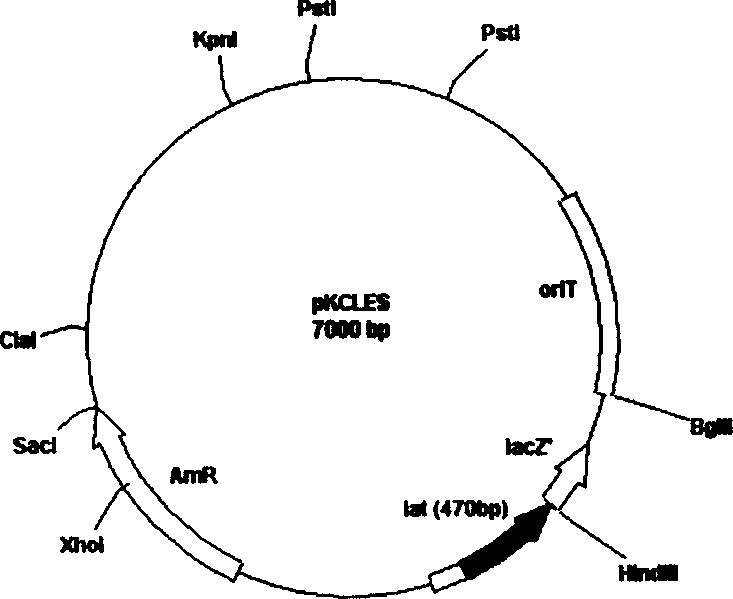
[0055] Construction of recombinant plasmid pKCLES
[0056] For the specific method, see ①, ②, ③, and ④ four steps in Example 1, and the ⑤ step is different from the restriction endonuclease used in the ⑤ step in Example 1, as follows:
[0057] The resulting recombinant plasmid pKC1139-lat was subjected to EcoRI-ScaI double enzyme digestion, a section of EcoRI-ScaI gene fragment with a size of about 1.4kb was cut out of the lat gene, and about 7.0kb linear pKCLES fragment was reclaimed (DV805A glue recovery kit of Takara Company), Use T for both cohesive ends 4 DNA polymerase (product of Takara company) fill level, fill 805A gel recovery kit), use T for its two sticky ends 4 DNA polymerase (product of Takara Company) was used to make up the level, and the leveling system was as follows: sterilized double-distilled water: 3.5 μL, T4 DNA polymerase buffer (10×): 1 μL, 0.1% BSA: 1 μL, recovered pKCLES: 2.5 μL, dNTP (2mM): 1μL, the total system of filling up reaction: 9μL; the ...
Embodiment 3
[0061] Derivative 1 of Escherichia coli-Streptomyces shuttle plasmid containing partial deletion of Streptomyces clavulicularis lat gene and its construction method
[0062] For the specific method, see ①, ②, ③, and ④ four steps in Example 1, the ⑤ step is different from the restriction endonuclease used in the ⑤ step in Example 1, and the specific steps are as follows:
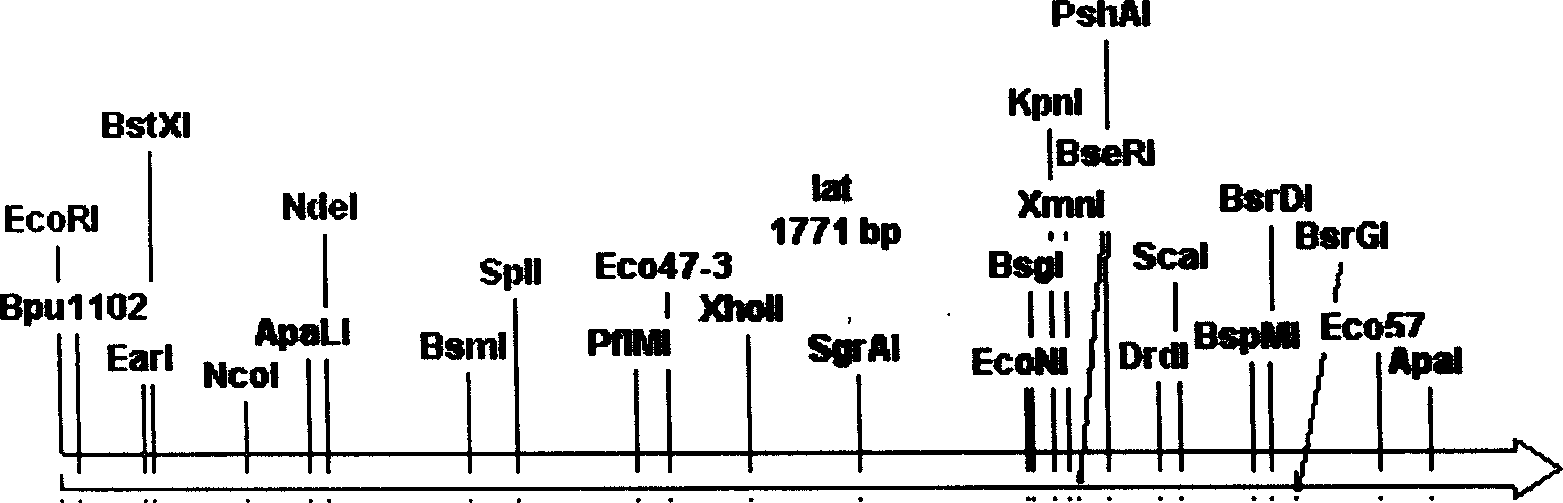
[0063] Adopt the rest of the enzyme cutting sites on the lat gene except HindIII-ScaI and EcoRI-ScaI used for constructing plasmid pKCLHS and pKCLES (see image 3 Any two enzymes in ), such as using EcoNI-HindIII to cut the lat gene in the plasmid pKC1139-lat, cut off a EcoNI-HindIII gene fragment with a size of about 0.7kb of the lat gene, and reclaim about a 7.6kb linear fragment, The two sticky ends were filled with T4 DNA polymerase (product of Takara Company), and the filling system was as follows: sterilized double-distilled water: 3.5 μL, T4 DNA polymerase buffer (10×): 1 μL, 0.1% BSA: 1 μL , DNA frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com