A set of oligonucleotide probe for detecting intestinal hemorrhage type colibacillus and vibrio cholerae
A technology of oligonucleotide probe and Escherichia coli, applied in the field of oligonucleotide probe and microbial detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Detect the screening and optimization of enterohaemorrhagic Escherichia coli O157:H7 and Vibrio cholerae O139 oligonucleotide probes
[0030] 1. Design of oligonucleotide probes for detection of enterohemorrhagic Escherichia coli O157:H7 and Vibrio cholerae O139
[0031]According to the Shiga-like toxin-producing genes stx1, stx2 and the β-glucuronidase gene (uidA) of enterohaemorrhagic Escherichia coli O157:H7; enterotoxin A subunit (ctxA) of Vibrio cholerae O139, virulence coordination 60 oligonucleotide probes were designed for pili A subunit (tcpA) and glycosyltransferase (LPSgt) genes. The sequence is shown in Table 1-1). The probe length is between 19-60 bases. When the probe is synthesized, its 3' terminal amino group is modified, which is used for the probe to react with the active aldehyde group on the surface of the slide and fix it to the surface of the carrier.
[0032] Table 1-1: Alternative sequences of oligonucleotide probes for detection of...
Embodiment 2
[0123] Example 2. Specificity and Sensitivity Evaluation of Oligonucleotide Probes
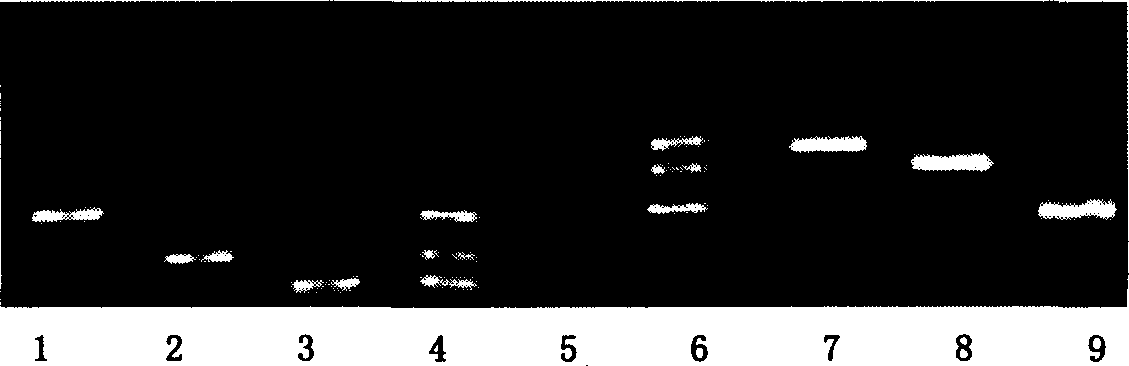
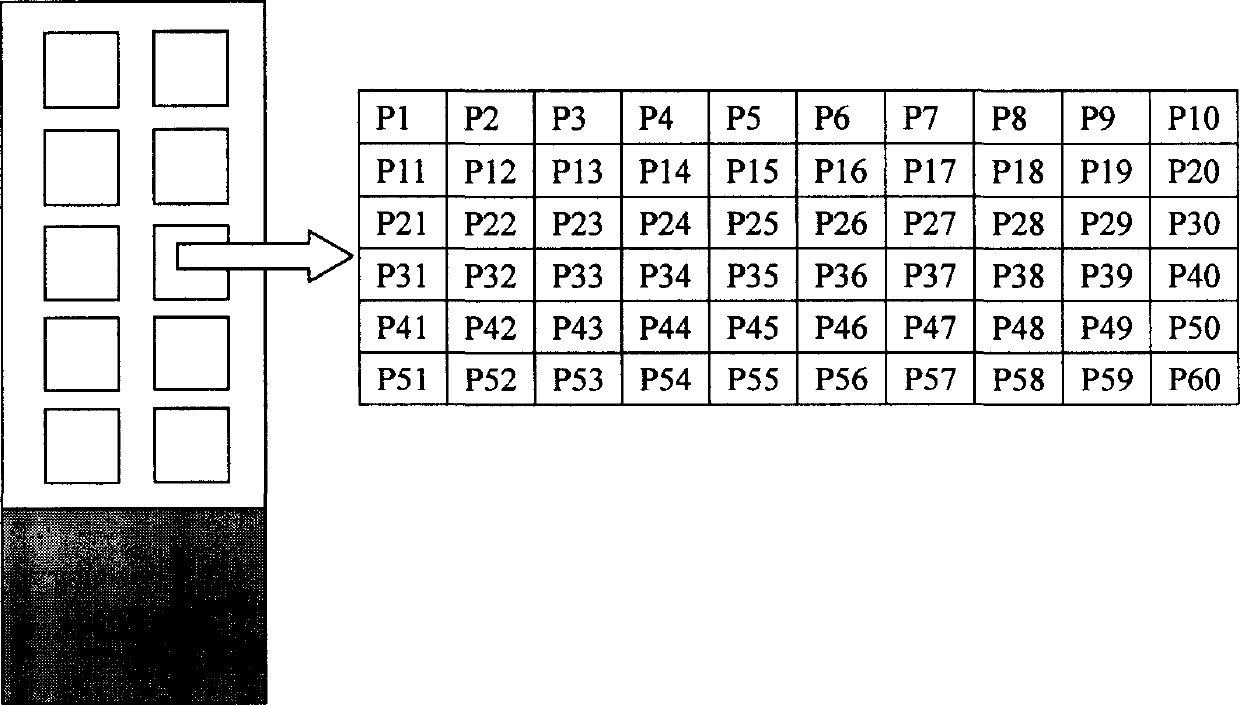
[0124] 1. Gene Chip Preparation The oligonucleotide probes were diluted with spotting solution (6×SSC, 0.1% SDS) to a final concentration of 50 μmol / L, and 5 ul was transferred to a 384-well plate. Use the Cartesian chip preparation instrument to spot the probes on the aldehyde-based chip (see Figure 4 for the arrangement of the probes, see Figure 5 for the hybridization results of the chip with fluorescently labeled triple PCR products of E. coli O157:H7 and Vibrio cholerae O139 respectively. ). In the probe microarray, the reverse complementary sequence of the fluorescent primer is used as a positive control, the irrelevant random sequence is used as a negative probe control, and the blank sample solution without probe is used as a negative control. The probe sequences are shown in Table 2-1.
[0125] Table 2-1 Gene chip probe sequences for detection of enterohemorrhagic Escherichia coli O...
Embodiment 3
[0149] Example 3. Detection of EHEC O157:H7 and Vibrio cholerae O139 oligonucleotide chip application
[0150] 1. Clinical sample collection and DNA extraction Take anal swabs from clinical patients at different stages of onset and treatment. Each sample was divided into two parts, one was routinely tested for bacteriology with traditional bacterial culture methods, and a total of 89 samples were detected as Vibrio cholerae 0139 (see Table 3-1 for the results); the other was grown in alkaline peptone water. Bacteria for 2-3 hours, then boiled and lysed for 10 minutes, centrifuged at 12,000 rpm for 2 minutes, and the DNA in the supernatant was taken as a template for multiplex PCR amplification.
[0151] 2. Oligonucleotide chip detection Escherichia coli O157:H7 and Vibrio cholerae O139 oligonucleotide chips were used to detect 342 clinical samples, and the clinical samples were subjected to multiple asymmetric PCR amplification with fluorescent primers, and the products were m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com