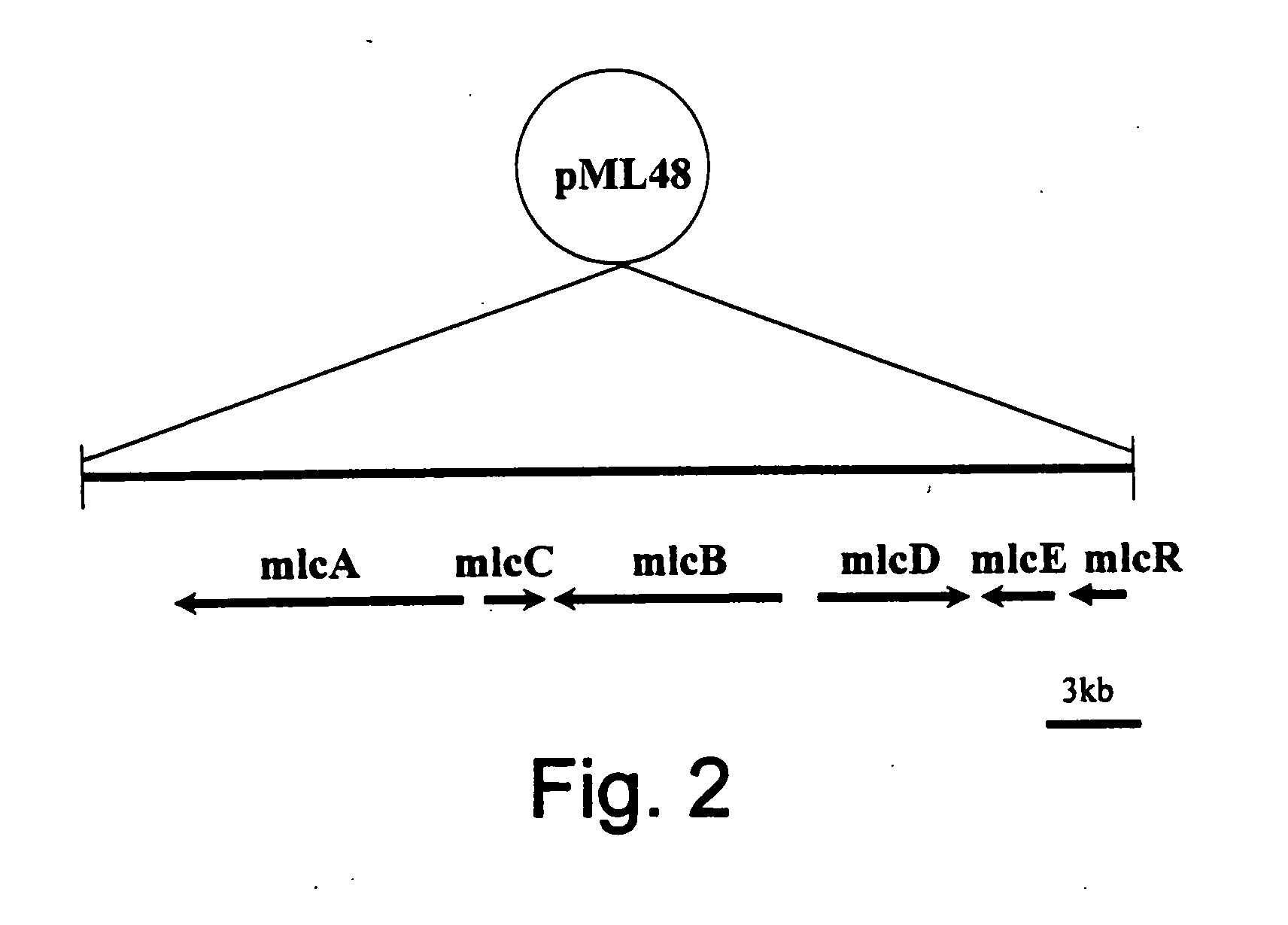
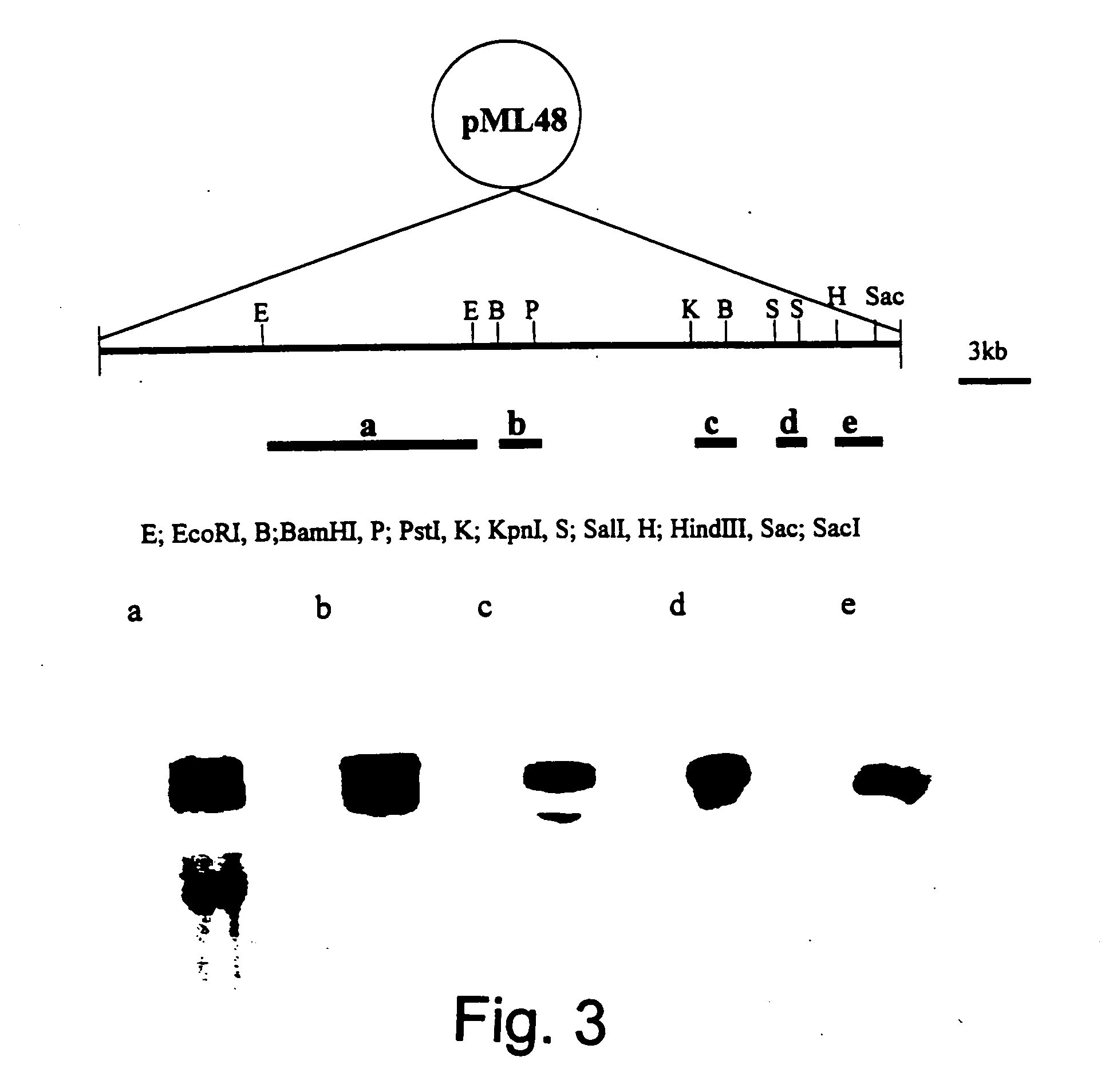
Genes from a gene cluster
a gene cluster and gene technology, applied in the field of gene clusters, can solve the problems of insufficient molecular biological analysis into the biosynthesis of ml-236b, and factors regulating
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
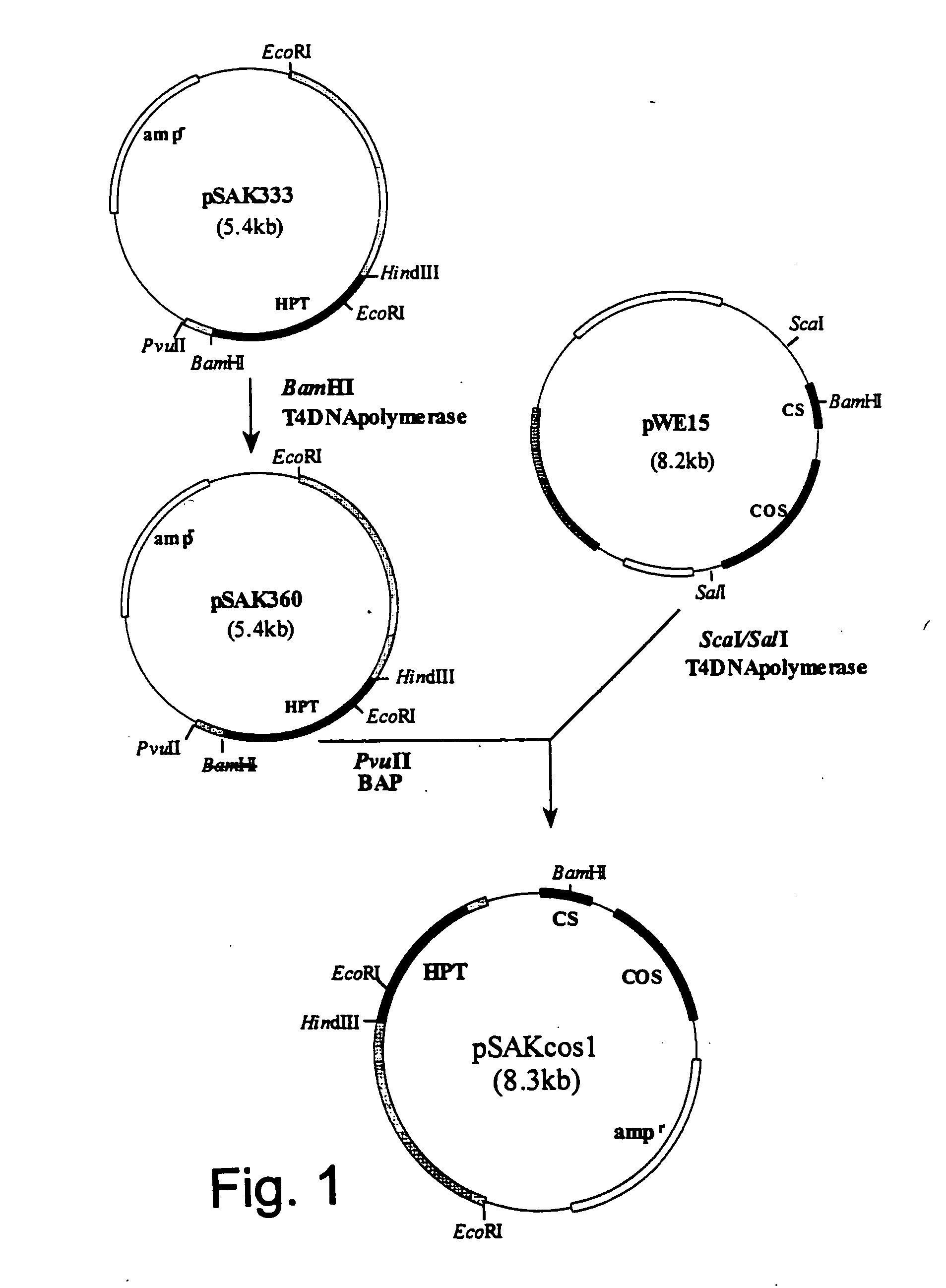
Construction of pSAKcos1 Vector
[0165] Plasmid pSAK333 containing the hygromycin B phosphotransferase gene (hereinafter referred to as “HPT”) originating from Escherichia coli (Japanese Patent Application Publication No. 3-262486) was digested with restriction enzyme BamHI (manufactured by Takara Shuzo Co., Ltd., Japan), and was treated to form blunt ends with T4DNA polymerase (manufactured by Takara Shuzo Co., Ltd., Japan).
[0166] The DNA fragment obtained as above was self-ligated into a circular form using DNA ligation kit Ver.2 (manufactured by Takara Shuzo Co., Ltd., Japan), and competent cells JM 109 of Escherichia coli (manufactured by Takara Shuzo Co., Ltd., Japan) were then transformed therewith. A strain having a plasmid in which the BamHI site was deleted was selected from the transformed Escherichia coli, and was designated pSAK360.
[0167] pSAK 360 was digested with restriction enzyme PvuII, and then treated with alkaline phosphatase to produce a fragment dephosphorylate...
example 2
Preparation of Genomic DNA of Penicillium citrinum SANK 13380
1) Culture of Penicillium citrinum SANK 13380
[0170] A seed culture of Penicillium citrinum SANK 13380 was made on a slant of PGA agar medium. Namely, the agar was inoculated with Penicillium citrinum SANK 13380 using a platinum needle, and kept at 26° C. for 14 days. The slant was kept at 4° C.
[0171] Main culturing was performed by liquid aeration culture. Cells from a 5 mm square of the above-mentioned slant were inoculated in 50 ml of MBG3-8 medium in 500 ml conical flask, and incubated at 26° C. with shaking at 210 rpm for five days.
2) Preparation of Genomic DNA from Penicillium citrinum SANK 13380
[0172] The culture obtained in step 1) was centrifuged at 10000×G at room temperature for 10 minutes, and cells were harvested. 3 g (wet weight) of cells were broken in a mortar cooled with dry ice so as to be in the form of a powder. The broken cells were put in a centrifuge tube filled with 20 ml of 62.5 mM EDTA 2Na (...
example 3
Preparation of Genomic DNA Library of Penicillium citrinum SANK13380
1) Preparation of Genomic DNA Fragment
[0176] 0.25 units of Sau3A1 (Takara Shuzo Co., Ltd., Japan) were added to 100 μl of an aqueous solution of genomic DNA (50 μg) of Penicillium citrinum SANK13380 obtained in Example 2. After intervals of 10, 30, 60, 90 and 120 seconds, 20 μl samples of the mixture were taken, and 0.5 M EDTA (pH 8.0) was added to each sample to terminate the restriction enzyme reaction. The resulting partially digested DNA fragments were separated by agarose gel electrophoresis, and agarose gel was recovered containing DNA fragments of 30 kb or more
[0177] The recovered gel was finely crushed, and placed into Ultra Free C3 Centrifuged Filtration Unit (manufactured by Japan Milipore corporation). The gel was cooled at −80° C. for 15 minutes until frozen, and then the gel was melted by incubating it at 37° C. for 10 minutes. It was centrifuged at 5000×G for 5 minutes, to extract DNA. The DNA was ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com