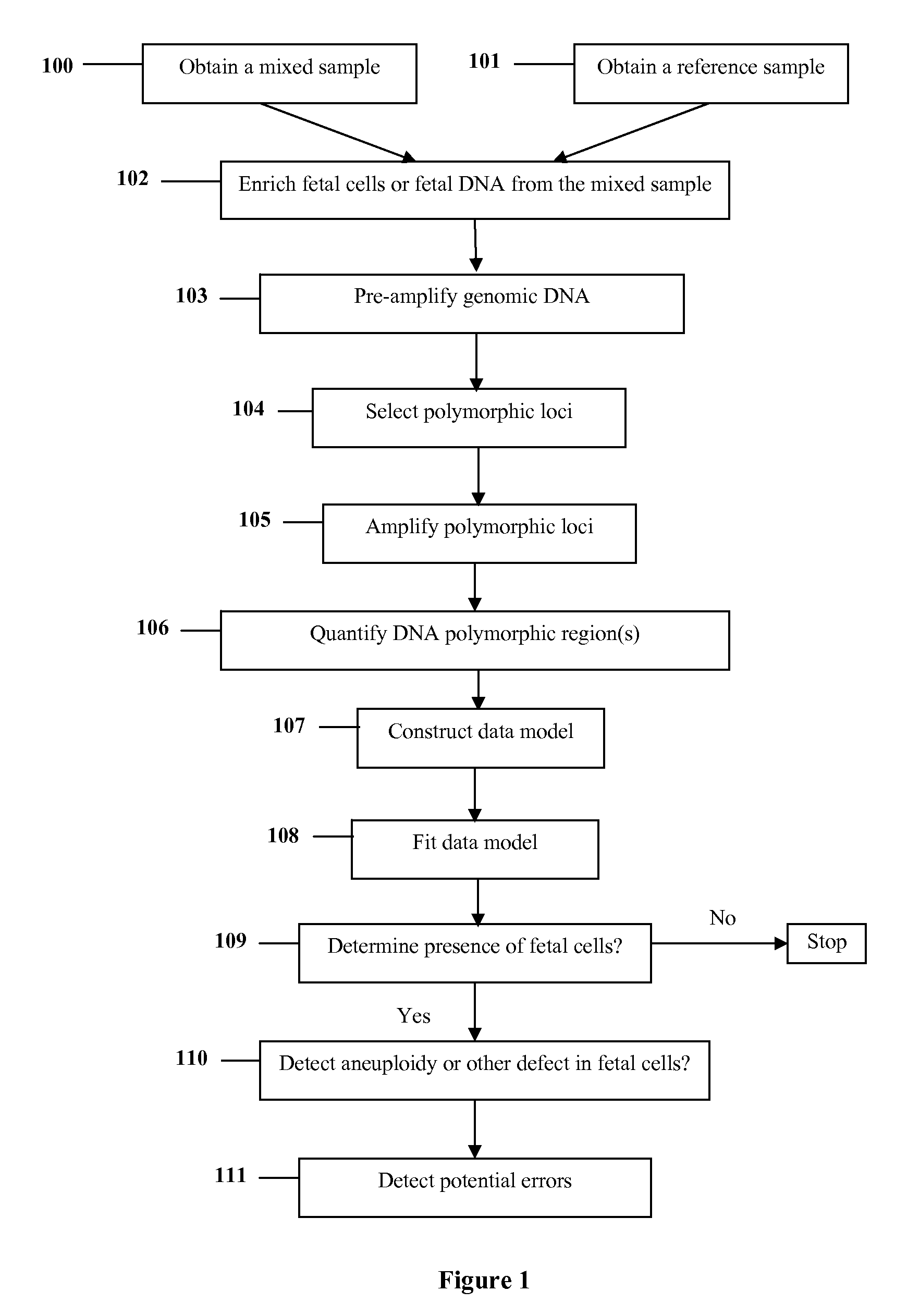
Diagnosis of fetal abnormalities using polymorphisms including short tandem repeats
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Separation of Fetal Cord Blood
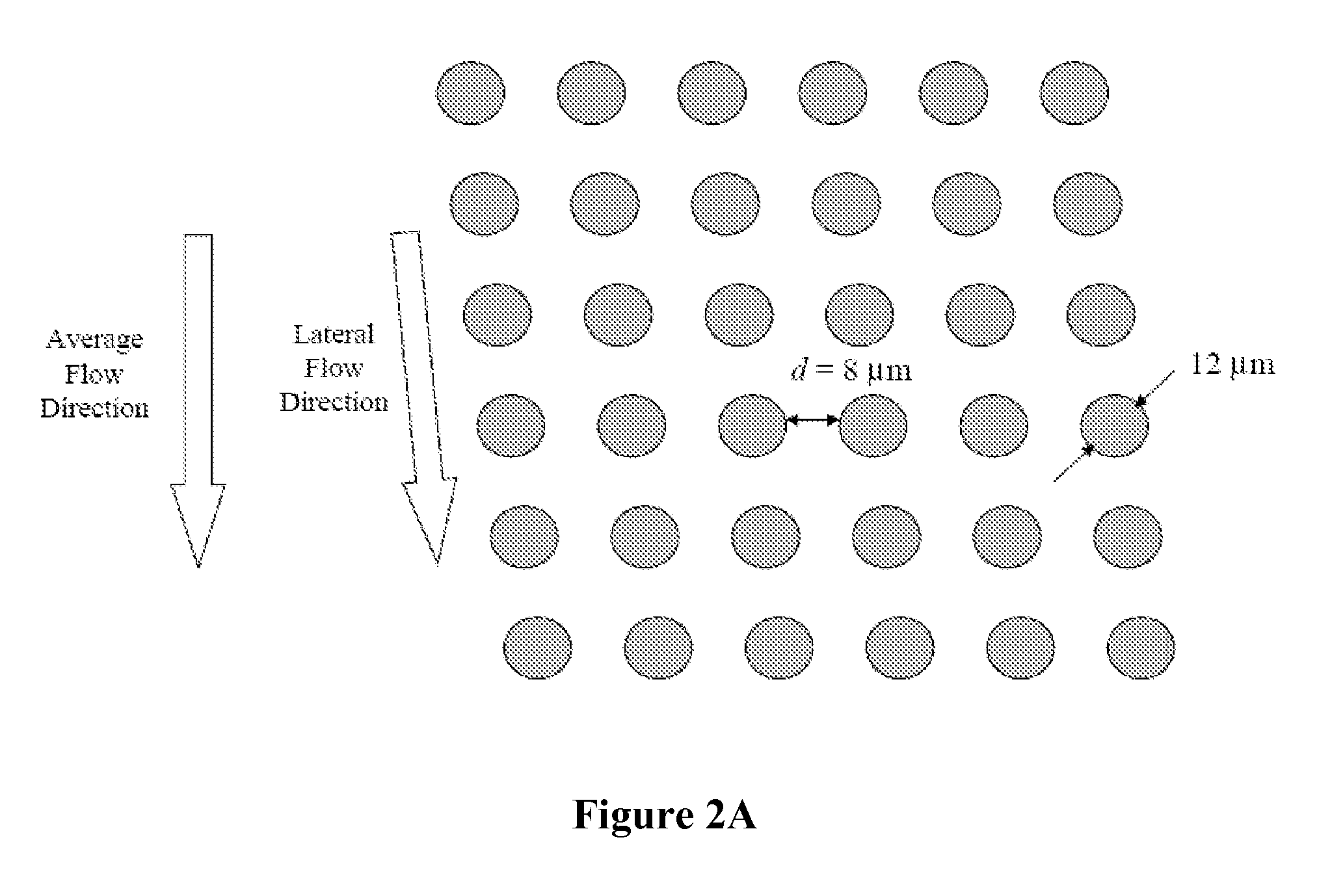
[0148]FIGS. 7A-7D illustrates a schematic of the device used to separate nucleated cells from fetal cord blood.
[0149] Dimensions: 100 mm×28 mm×1 mm
[0150] Array design: 3 stages, gap size=18, 12 and 8 μm for the first, second and third stage, respectively.
[0151] Device fabrication: The arrays and channels were fabricated in silicon using standard photolithography and deep silicon reactive etching techniques. The etch depth is 140 μm. Through holes for fluid access are made using KOH wet etching. The silicon substrate was sealed on the etched face to form enclosed fluidic channels using a blood compatible pressure sensitive adhesive (9795, 3M, St Paul, Minn.).
[0152] Device packaging: The device was mechanically mated to a plastic manifold with external fluidic reservoirs to deliver blood and buffer to the device and extract the generated fractions.
[0153] Device operation: An external pressure source was used to apply a pressure of 2.0 PSI to the buf...
example 2
Isolation of Fetal Cells from Maternal Blood
[0157] The device and process described in detail in Example 1 were used in combination with immunomagnetic affinity enrichment techniques to demonstrate the feasibility of isolating fetal cells from maternal blood.
[0158] Experimental conditions: blood from consenting maternal donors carrying male fetuses was collected into K2EDTA vacutainers (366643, Becton Dickinson, Franklin Lakes, N.J.) immediately following elective termination of pregnancy. The undiluted blood was processed using the device described in Example 1 at room temperature and within 9 hrs of draw. Nucleated cells from the blood were separated from enucleated cells (red blood cells and platelets), and plasma delivered into a buffer stream of calcium and magnesium-free Dulbecco's Phosphate Buffered Saline (14190-144, Invitrogen, Carlsbad, Calif.) containing 1% Bovine Serum Albumin (BSA) (A8412-100 mL, Sigma-Aldrich, St Louis, Mo.). Subsequently, the nucleated cell fraction...
example 3
Confirmation of the Presence of Male Fetal Cells in Enriched Samples
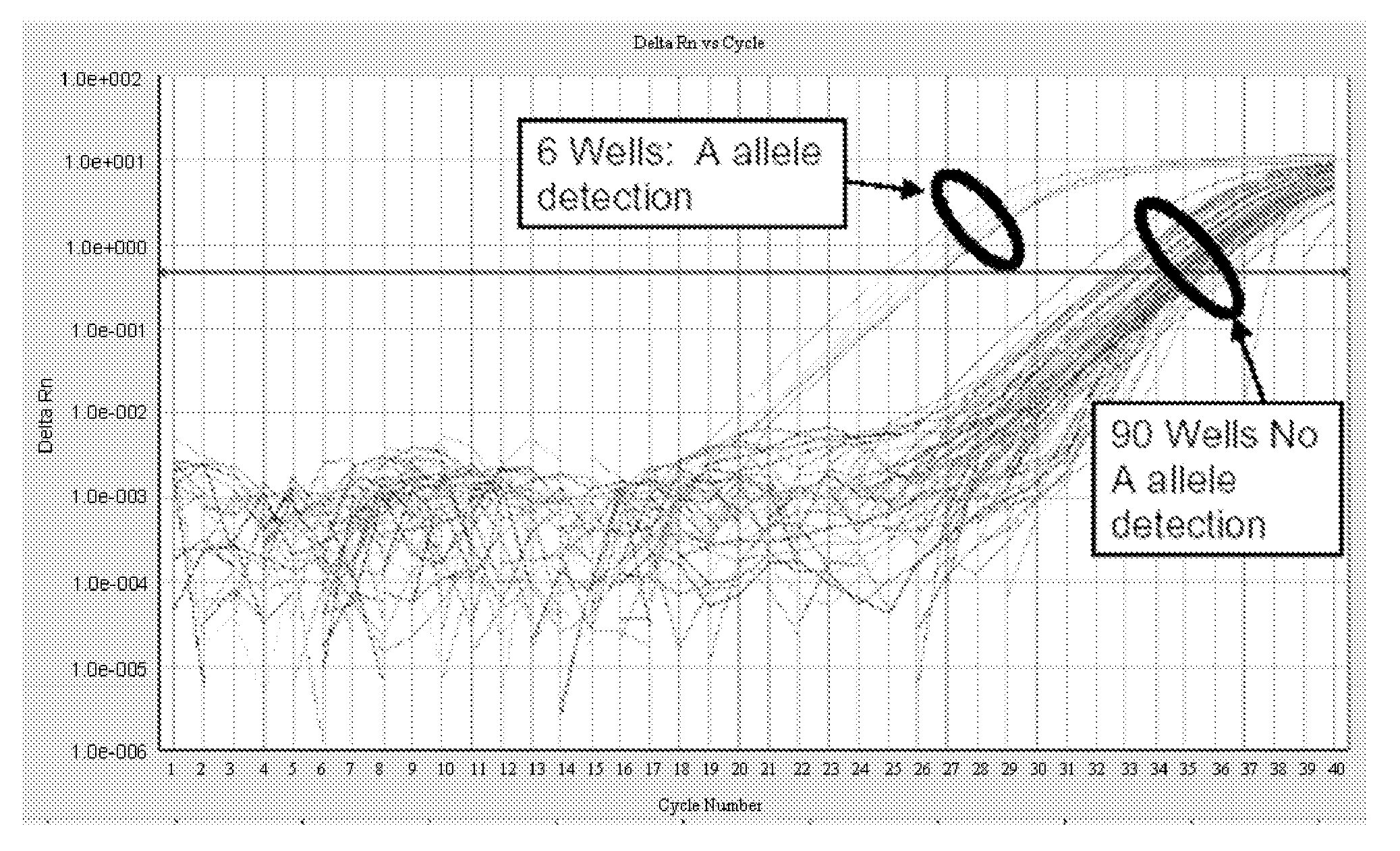
[0161] Confirmation of the presence of a male fetal cell in an enriched sample is performed using qPCR with primers specific for DYZ, a marker repeated in high copy number on the Y chromosome. After enrichment of fnRBC by any of the methods described herein, the resulting enriched fnRBC are binned by dividing the sample into 100 PCR wells. Prior to binning, enriched samples may be screened by FISH to determine the presence of any fnRBC containing an aneuploidy of interest. Because of the low number of fnRBC in maternal blood, only a portion of the wells will contain a single fnRBC (the other wells are expected to be negative for fnRBC). The cells are fixed in 2% Paraformaldehyde and stored at 4° C. Cells in each bin are pelleted and resuspended in 5 μl PBS plus 1 μl 20 mg / ml Proteinase K (Sigma #P-2308). Cells are lysed by incubation at 65° C. for 60 minutes followed by inactivation of the Proteinase K by incubatio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com