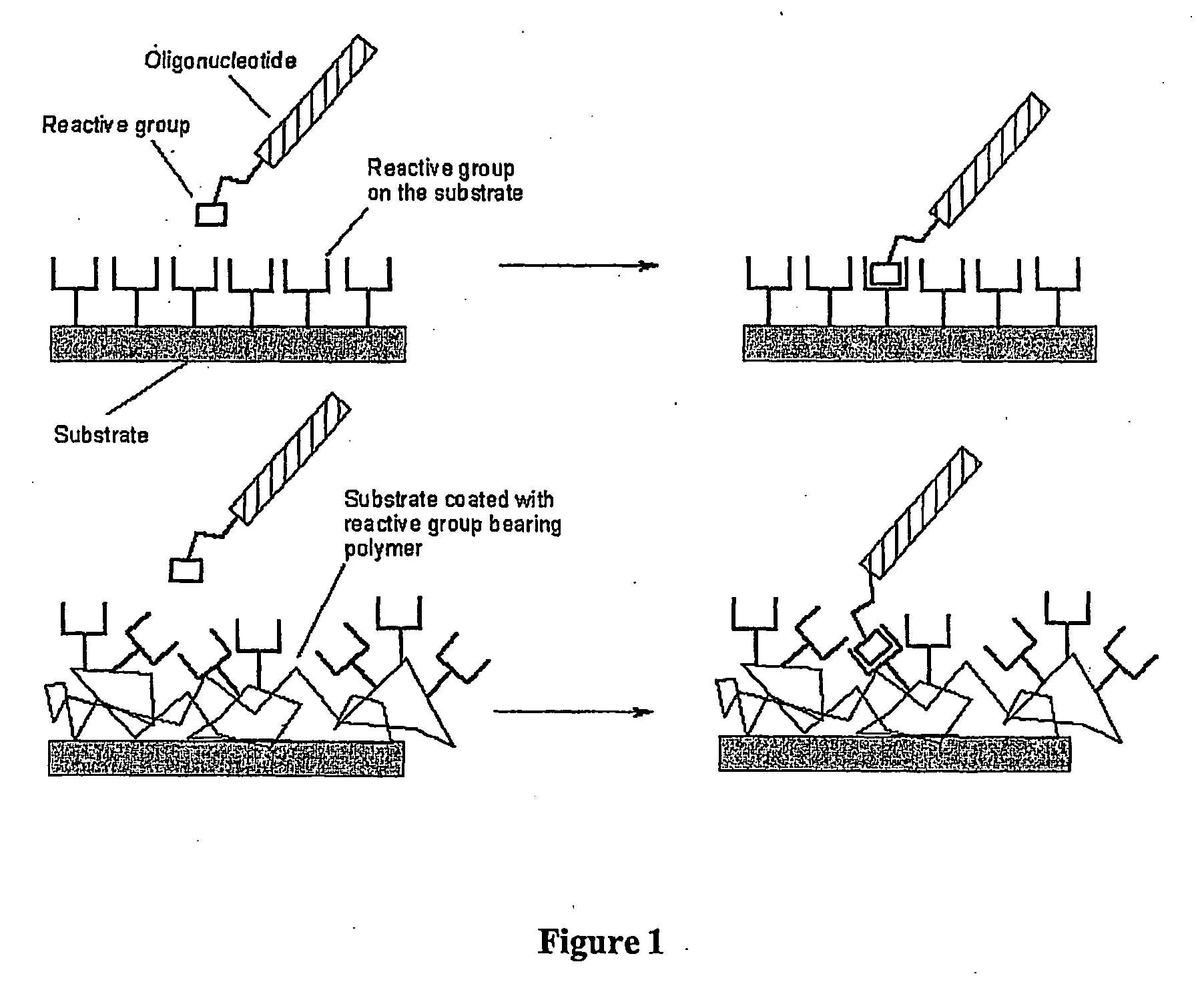
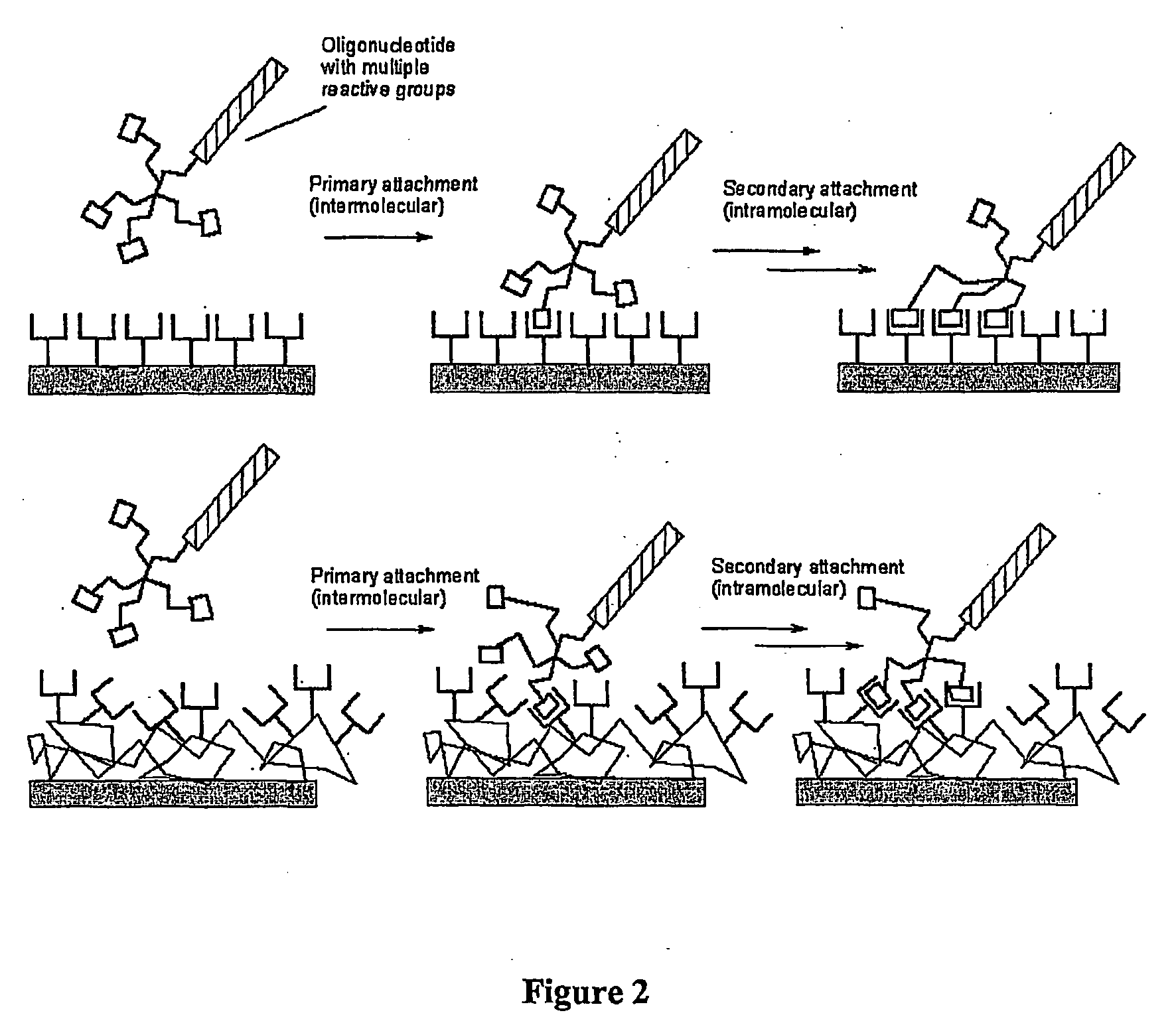
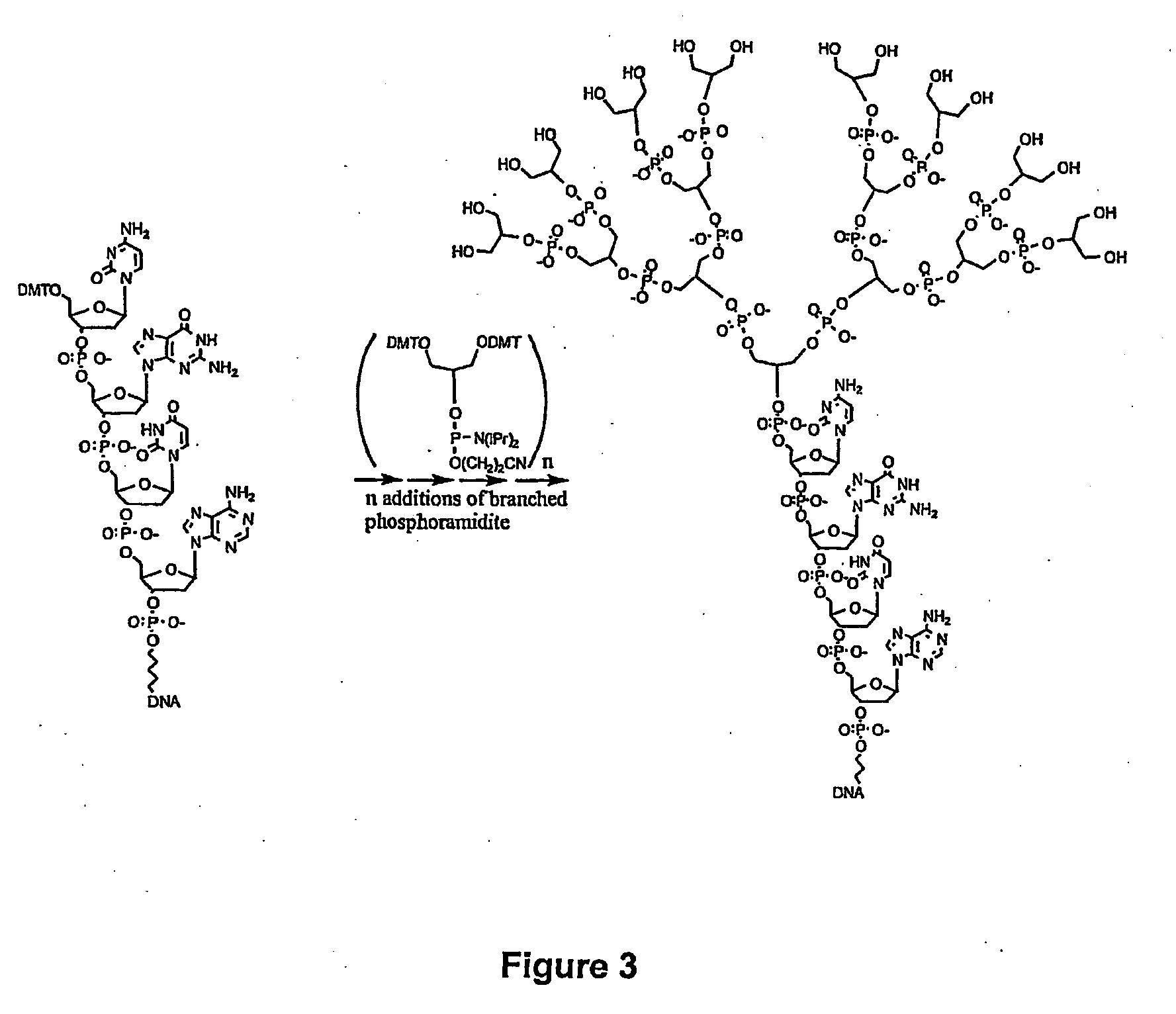
Biomolecules having multiple attachment moieties for binding to a substrate surface
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Experiment 1.1
Synthesis of N-Triphenylmethyl-6-hydroxycapronic acid hydrazide, (compound 5, FIG. 9A)
[0064]To a solution of 6.2 g (20 mmol) of tritylhydrazine hydrochloride (3a) in 200 ml of THF was added 2.22 g (22 mmol, 1.1 eq) triethylamine. The solution was stirred at room temperature (rt) for 15 min, filtered, concentrated to afford compound 3, then treated with 2.29 g (20 mmol, 1 eq) of ε-caprolactone (compound 4). The mixture is heated to 65° C. for 5 h the cooled to rt for 18 h. The precipitate was collected and recrystallized from ethyl acetate to afford 3.55 g (45%) of a white powder (compound 5): 1H-NMR 7.49-7.47 (m, 5H), 7.35-7.10 (m, 10H), 6.55 (d, J=7.52, 1H), 5.55 (d, J=7.25, 1H), 3.54 (t, J=6.45, 2H), 1.87 (t, J=7.25, 2H), 1.62 (bs, 1H), 1.57-1.34 (m, 4H), 1.27-1.11 (m, 2H).
experiment 1.2
Synthesis of 6-[(2Cyanoethoxy)(diisopropylamino)phosphanyloxy]-N′-tritylhexanohydrazide (compound 1a, FIG. 9A)
[0065]To a solution of 3.0 g (7.7 mmol) N-triphenylmethyl-6-hydroxycapronic hydrazide (compound 5) in 50 ml dry dichloromethane at rt was slowly added 4.0 g (31 mmol, 4 eq) of N-ethyldiisopropyl amine and 2.01 g (8.5 mmol, 1.1 eq) of chloro(diisopropylamino)-β-cyanoethoxyphosphine (compound 6) over 15 min. Upon complete addition, the reaction was stirred for 1 h, concentrated, and chromatographed (ethyl acetate / n-heptane ⅔ with 0.2% triethylamine) to afford 3.19 g (70%) of 1a as a pale yellow foam.
[0066]1H-NMR: 7.49-7.46 (m, 5H), 7.34-7.20 (m, 10H), 6.57 (d, J=7.2, 1H), 5.57 (d, J=7.5, 1H), 3.85-3.74 (m, 2H), 3.62-3.48 (m, 4H), 2.62-2.59 (m, 2 H), 1.88-1.84 (m, 2H), 1.53-1.33 (m, 4H), 1.27-1.13 (m, 14H); 31P-NMR (CDCl3): δ=147.97.
experiment 1.3
Preparation of Ethyl 6-[(2-cyanoethoxy)(diisoprooylamino)phosphanyloxy]hexanoate (compound 1b, FIG. 9B. Scheme 2)
[0067]To a solution of 1.65 g (10 mmol) of ethyl 6-hydroxyhexanoate (compound 7) in 30 ml dichloromethane at rt are slowly added 5.17 g (40 mmol, 4 eq) of N-ethyldiisopropyl amine and 2.6 g (11 mmol, 1.1 eq) of compound 6 over 15 min. Upon complete addition, the reaction was further stirred for 15 min, concentrated, and chromatographed (ethyl acetate / n-heptane ¼ with 0.2% triethylamine) to afford 2.47 g (69%) of compound 1b as clear oil: 1H-NMR 4.12 (q, J=7.25, 2H), 3.90-3.77 (m, 2H), 3.75-3.55 (m, 4H), 2.64 (t, J=6.44, 2H), 2.30 (t, J=7.25, 2H), 1.69-1.59 (m, 4H), 1.44-1.34 (m, 2H), 1.25 (t, J=7.25, 3H), 1.20-1.12 (m, 12H); 31P-NMR (CDCl3): δ=148.01.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Structure | aaaaa | aaaaa |
| Covalent bond | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com