Antibody production
a technology of antibody and vh gene, applied in the field of antibody production, can solve the problems of non-productive response to antigen challenge, limited expression of transgenes across target tissues, and remains technically demanding, and achieves the effects of enhancing the probability of vh region, and enhancing the probability of all vh gene segments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
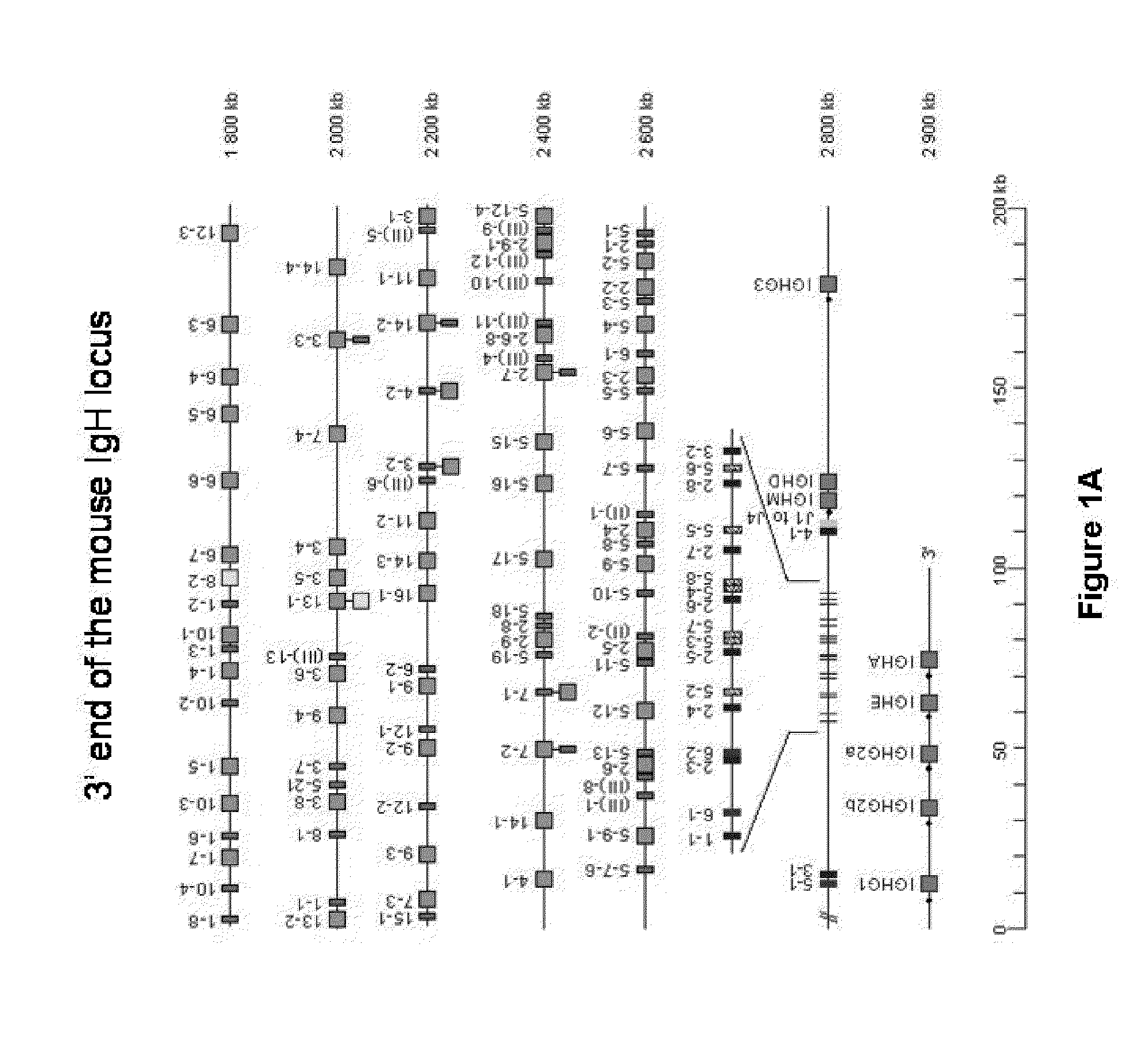
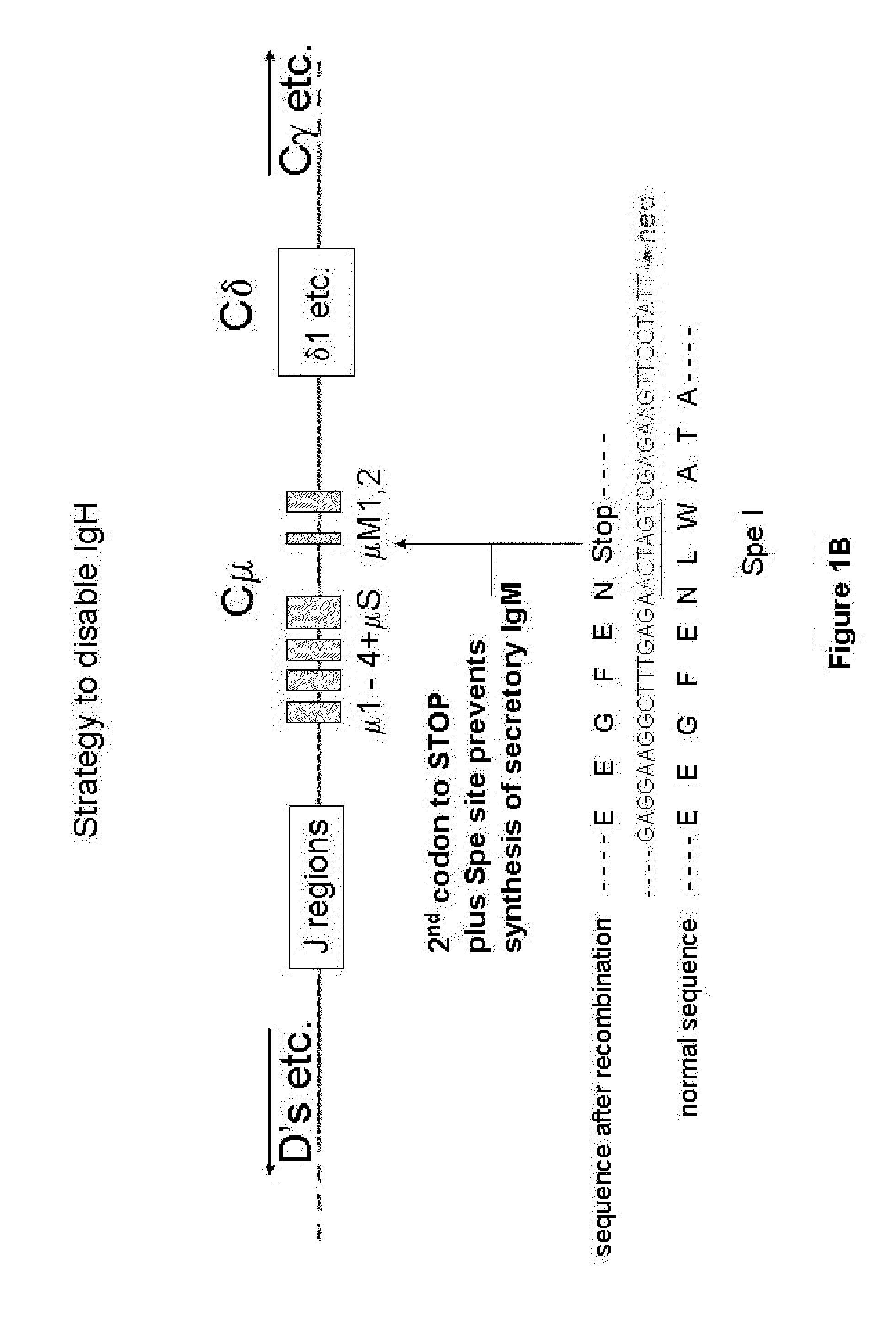
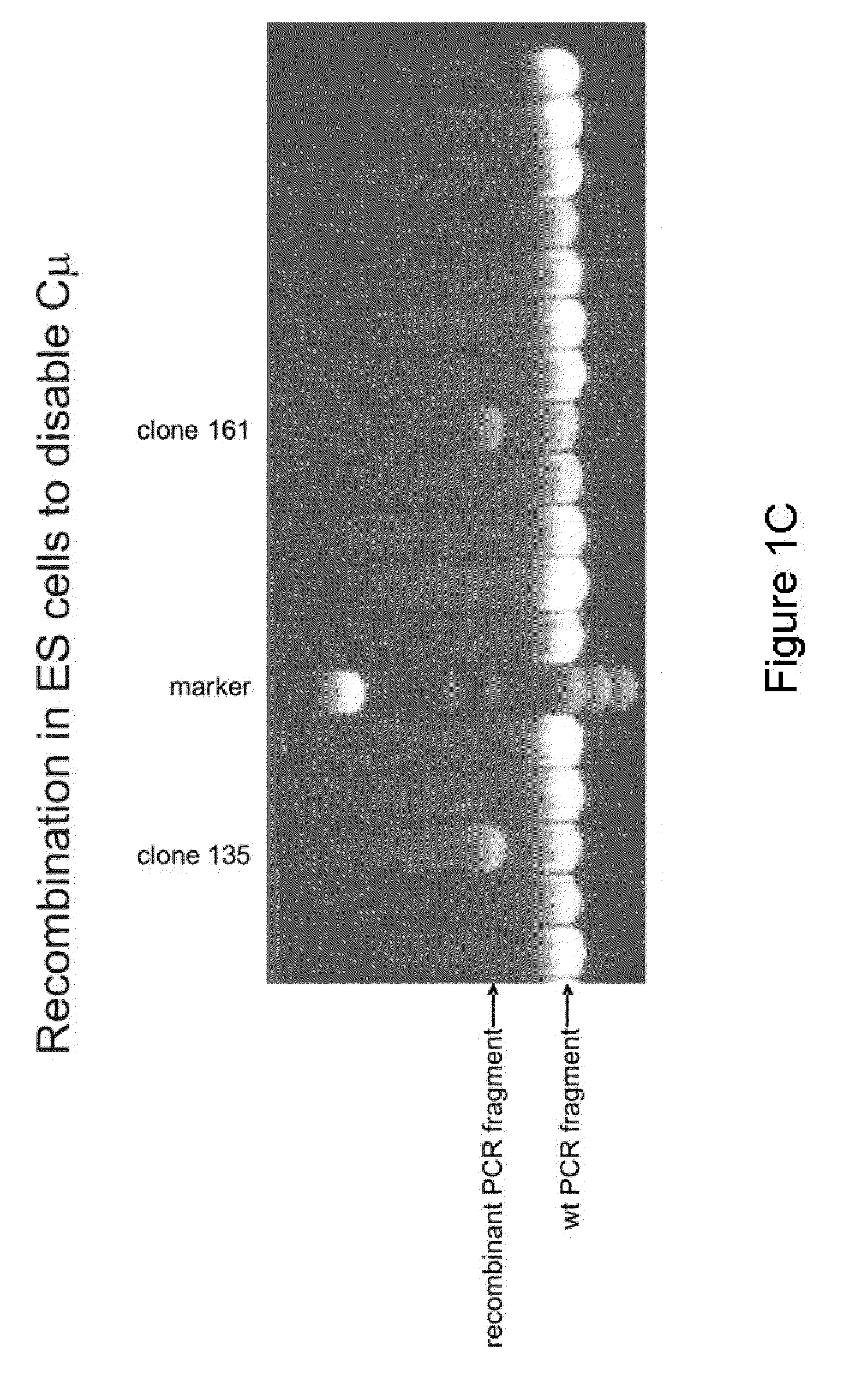
[0157]In this example, the IgH locus (FIG. 1A) is inactivated by a strategy similar to that published by Kitamura and Rajewsky with the difference that the stop codon is introduced into the Cμ regions at a position one amino acid before that described by Kitamura et al. (1991) Nature, 350, 423-426. ES (or iPS) cells were transfected with a construct that changes the second codon of the first membrane exon of the mouse IgM gene into a stop codon. This involves routine procedure including a neo selection for transfection. A SpeI site was included in the recombination sequences to be able to monitor the successful recombination (FIG. 1B). ES cells are subsequently screened by Southern blots to confirm successful recombinant clones. This resulted in 10 correct recombinants (e.g. FIG. 1C). Of these, 3 were injected into mouse blastocysts to obtain chimeras which were subsequently bred to obtain mice that are homozygous for the IgH mutation. FACS analysis (B220 versus CD19) of the B cells...
example 2
[0167]This example is in principle the same as Example 1 with the exception that the high frequency of obtaining IgH / Igκ H2L2 antibodies is increased even further by lowering the frequency of expression of the endogenous mouse Igλ locus. This can be achieved by replacing the regulatory regions of both Igλ with a selectable marker (FIG. 11), in this case the hygromycin resistance gene and the TK-BSD gene.
[0168]The latter allows positive selection, resistance to blasticidin S (Karreman, (1998) NAR, 26, (10), 2508-2510). This combination of markers allows for positive selection in the two ES cell recombinations when replacing the regulatory regions. The recombination would be carried out in the ES cells generated in Example 1 or alternatively in parallel in normal ES cells and bred into the mice described above in Example 1.
[0169]The resulting transgenic mice would contain the hybrid human rat IgH and Igκ loci, be negative for endogenous mouse IgH and Igκ (or express Cκ at very low lev...
example 3
[0170]Example 3 is analogous to the Examples described above but the hybrid Igκ locus would be extended by the addition of Vκ segments that are used less frequently (FIG. 12; Vκ 1-9, 1-33, 2-30, 2-28, 1-27, 1-5). Alternatively, mutated / modified Vκ segments or Vλ segments could be added in addition. The addition of further segments would be carried out by using the same XhoI / SalI cloning strategy described above. Immunization of mice generated in this example would allow a greater complexity in response to the immunization with antigen. The number of VL regions could be varied further by adding other Vκ segments or the use of combinations of all of the above VL segments.
PUM
| Property | Measurement | Unit |
|---|---|---|
| structure | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
| γ | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com