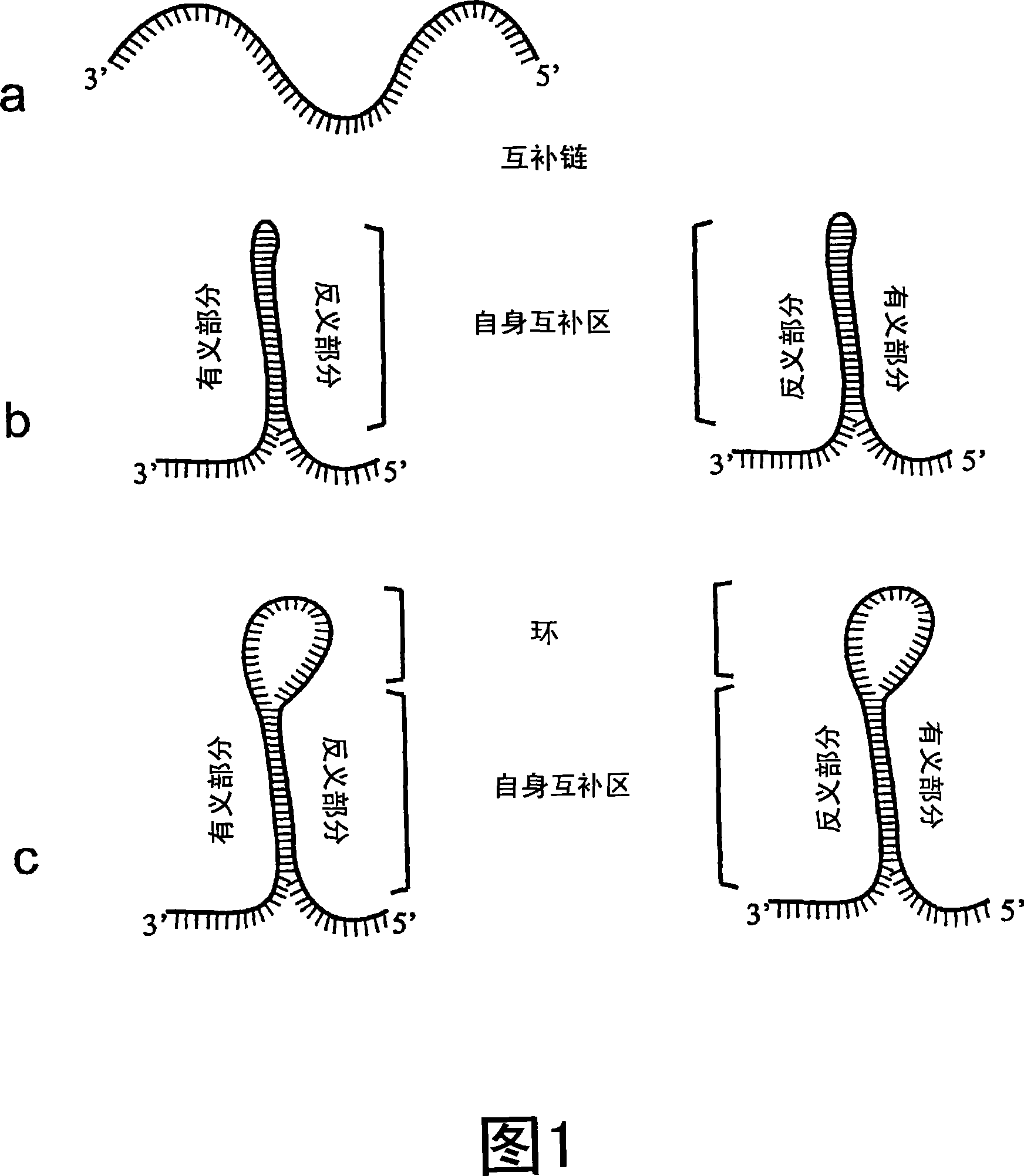
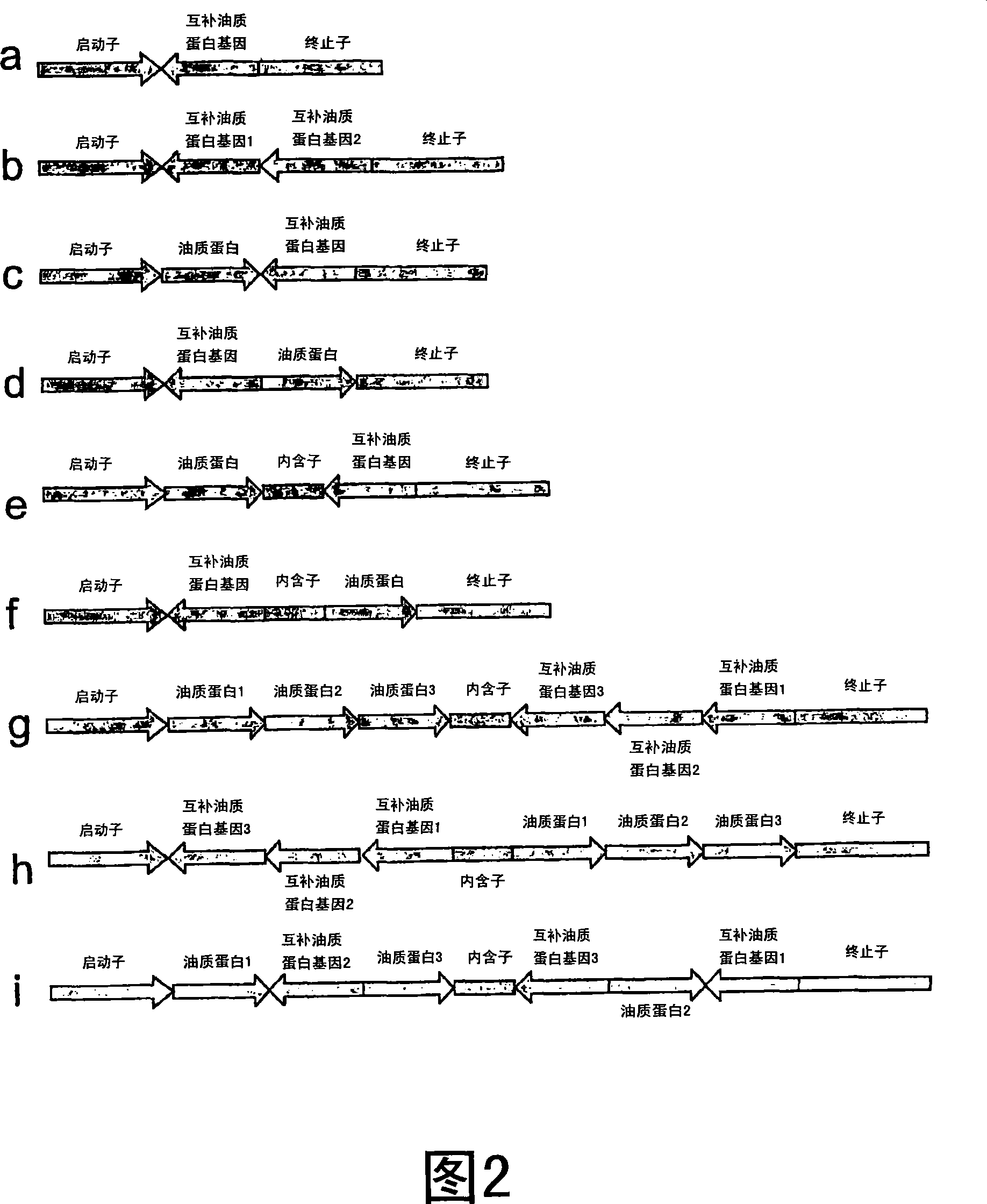
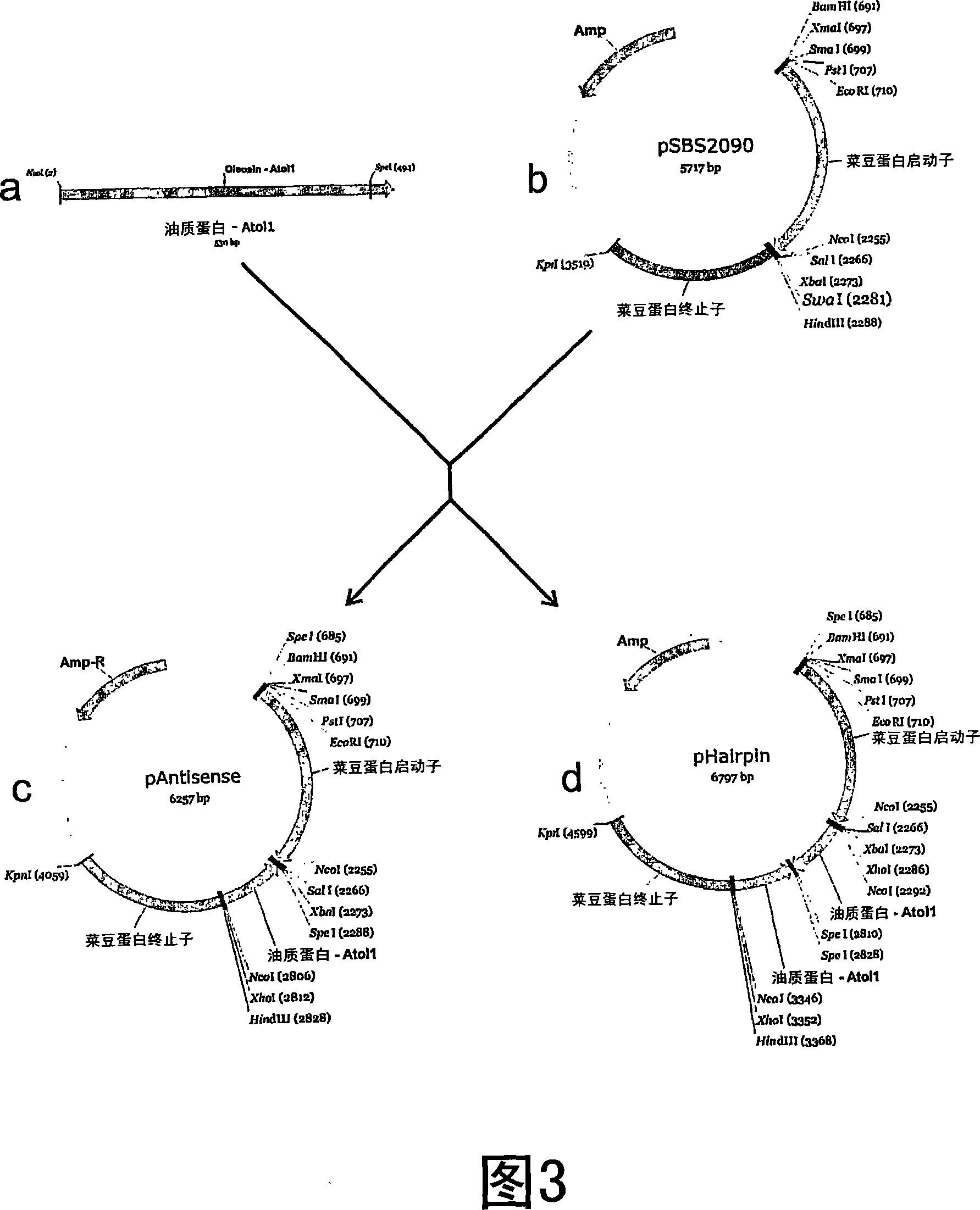
Methods for the modulation of oleosin expression in plants
A technology for oleosin and plants, applied in the field of plant genetic engineering construction, can solve the problems of not being able to specifically inhibit specific genes, unclear oleosin gene expression, and impracticality of T-DNA Agrobacterium insertion mutagenesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0269] Example 1: Construction of an oleosin suppression cassette
[0270] Antisense box
[0271] Use the forward primer NTD (5’-TATT) containing restriction sites HindIIII and NcoI (underlined) AAGCTTCCATGG CCGATACTGCTAGAGG-3') (SEQ ID NO: 92) and the reverse primer CTR (5'-AGCCAT) containing the restriction site SpeI (underlined) ACTAGT AGTGTGTTGACCACCACGAG-3') (SEQ ID NO: 93), and Atol1 cDNA (SEQ ID NO: 94) was used as a template to amplify Atol1 cDNA. The PCR product was purified and inserted into the vector pSBS2090 controlled by the phaseolin promoter / terminator (Slightom et al., 1983 Proc. Natl. Acad. Sci. U.S.A. 80:1897-1901). The vector was previously digested with restriction enzyme SwaI (Figure 3b). Insert the PCR product in cis or trans orientation because the enzyme SwaI can produce blunt ends. The plasmid containing Atol1 cDNA in trans orientation was screened by NcoI. The vector containing Atol1 cDNA in the trans-direction digested by NcoI released a 551 bp DNA fra...
Embodiment 2
[0277] Example 2: Transformation of Agrobacterium and Arabidopsis
[0278] By electroporation, the binary vectors pSBS3000-antisense sequence, pSBS3000-hairpin and pSBS3000-hairpin+intron were inserted into Agrobacterium EHA101 (Hood, EE et al., 1986. Journal of Bacteriology 168: 1291-1301). ). Using spectinomycin resistance ("SpecR" in Figure 5e), the transformed Agrobacterium line containing the binary vector was selected. One Agrobacterium line was selected for each construct.
[0279] The Arabidopsis ecotype C24 was used for transformation. In a 4-inch pot, five seeds were planted on the surface of the soil mixture (two-thirds of Redi soil and one-third of perlite, pH=6.7). Let the seedling grow to the 6-8 leaf rosette stage, about 2.5cm in diameter. These seedlings were transplanted into a 4-inch pot containing the soil mixture described above, and covered with a screen material with five meshes of 1 cm diameter, one in the corner and one in the center. The pot was placed on ...
Embodiment 3
[0283] Example 3: Separation of oil bodies
[0284] The accumulation of Atol1 in the seeds recovered from the selected strain was analyzed by SDS-PAGE of the oil body part. Using the method reported by van Rooijen Moloney ((1995) Biotechnology (N.Y.) 13, 72-77), the oil bodies of these seeds were obtained by the following modifications. Briefly, in a 1.7 ml microcentrifuge tube with 0.4 ml of oil body extraction buffer (50 mM Tris-HCl with 0.4 M sucrose and 0.5 M NaCl, pH 7.5), mill 10 to 20 mg of dry mature seeds. The extract was centrifuged at 10,000 g for 15 minutes at room temperature (RT). After centrifugation, the lipid layer containing oil bodies was removed from the water phase and transferred to another microcentrifuge tube. The oil body is suspended in 0.4ml of high stringency urea buffer (100mM sodium carbonate buffer containing 8M urea, pH8.0). The sample was centrifuged at 10,000 g for 15 minutes at 4°C, and the undernatant was removed. Finally, the oil bodies are sus...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com