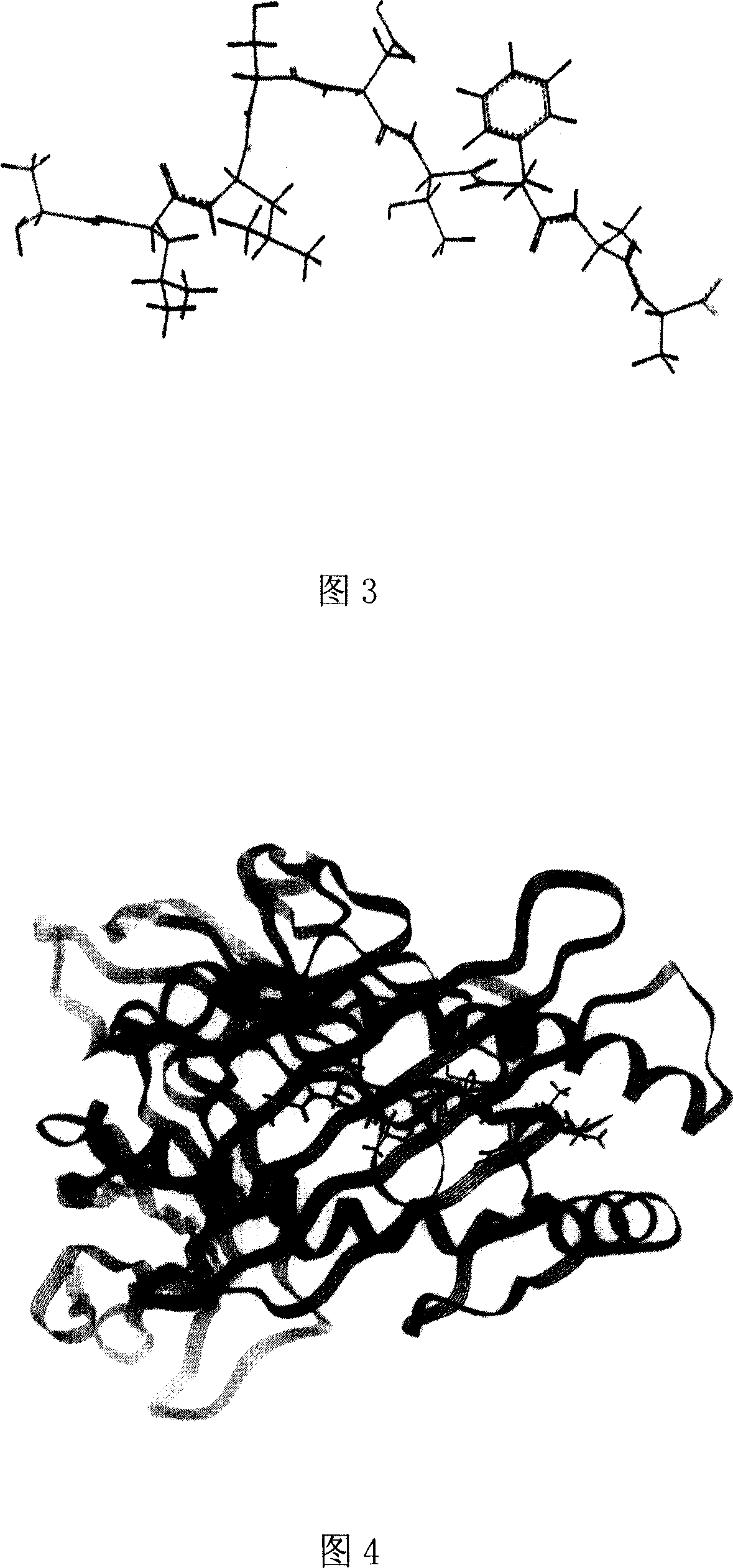
Heparinase polypeptide epitope combined to molecules in human MIIC -I category
A technology of heparinase and binding sites, applied in the direction of animal/human peptides, medical preparations containing active ingredients, peptides, etc., to achieve the effects of convenient clinical application, improved accuracy, and large commercial industrialization value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] The screening method of embodiment 1 polypeptide epitope
[0036] The polypeptide sequence obtained in the present invention is directly retrieved from the peptide library through the combination of the supermotif method and the quantitative motif method, and maintains the binding property of MHC-I molecules. Through probabilistic and statistical analysis of these polypeptide sequences, a binding model of the polypeptide and MHC-I class molecules is obtained.
[0037] 1. Obtaining the amino acid sequence of human heparanase
[0038]The full-length amino acid sequence (543 amino acids in total) of the natural human heparanase found in the international open shared gene bank NCBI GeneBank is expressed as: SEQ No.1: Met Leu Leu Arg Ser Lys Pro Ala Leu Pro Pro Pro ProLeu Met Leu Leu Leu Leu Gly Pro Leu Gly Pro Leu Ser Pro Gly Ala Leu Pro Arg ProAla Gin Ala Gin Asp Val Val Asp Leu Asp Phe Phe Thr Gin Glu Pro Leu His Leu ValSer Pro Ser Phe Leu Ser Val Thr Ile Asp Ala Asn Leu...
Embodiment 2
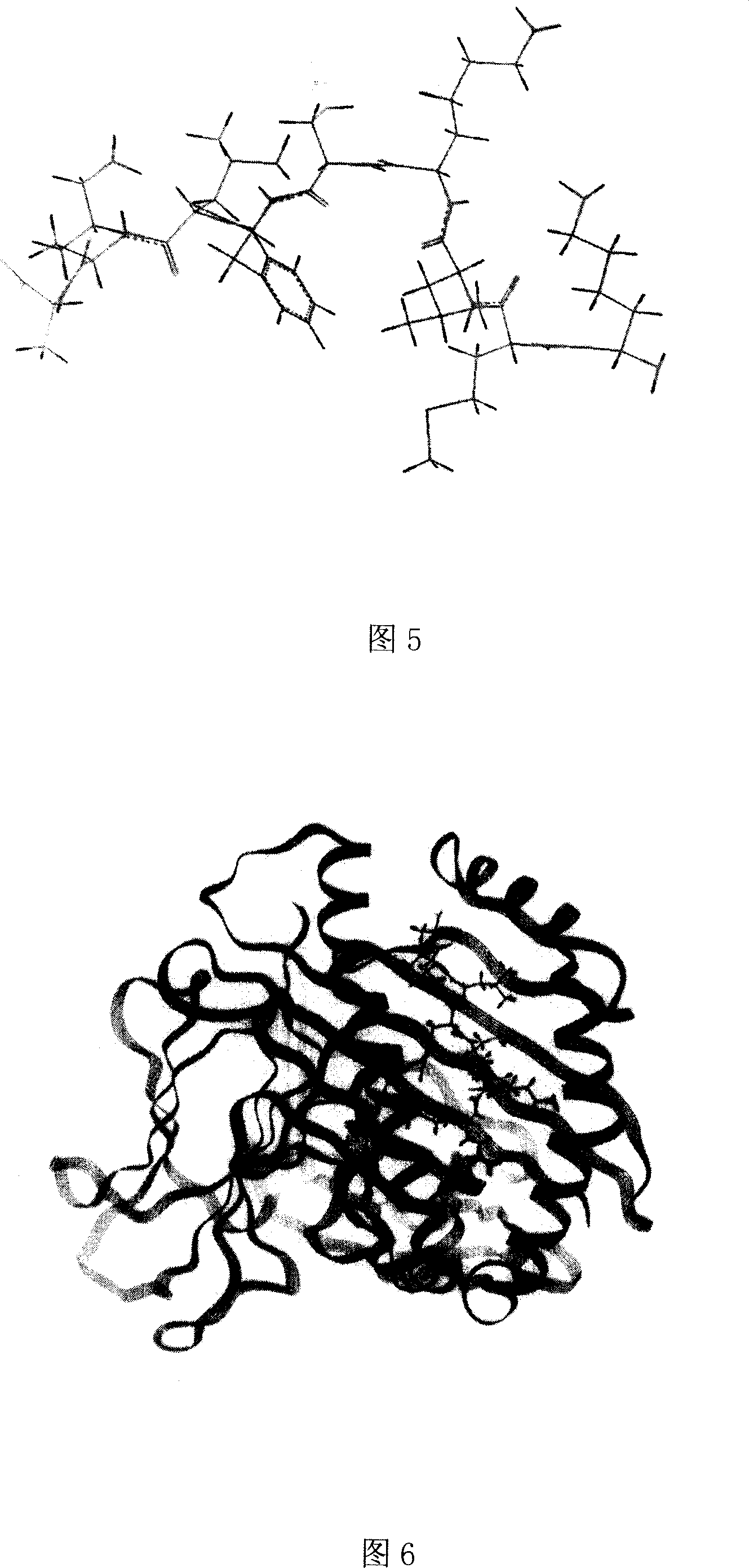
[0058] Example 2 Three-dimensional structure analysis and molecular dynamics simulation analysis of polypeptide epitope binding to human MHC-I
[0059] Using the Silicon graphics workstation and Insight11 software to establish the three-dimensional structure of the predicted polypeptide epitope binding to HLA-A2.1 and perform molecular dynamics simulation on the above-mentioned obtained polypeptide epitope, including the following methods: ①Molecular model construction, using the Insight II software package The Discover 3 module (using CVFF force field) performed molecular dynamics simulations on complexes of several nonapeptides from HPA with HLA-A2.1. The initial coordinates of HLA-A2.1 come from the complex of HLA-A0201 and influenza virus protein M1 (58-66) in the protein crystal structure database (Protein Data Bank entry1HHI). obtained by amino acid substitutions. The simulation process is as follows: first, HLA and β2m are fixed, and the nonapeptide is optimized for 20...
Embodiment 3
[0063] Obtaining of the polypeptide epitope of the present invention
[0064] (1) Synthesis of peptides
[0065] The synthesis of the peptide was carried out on the ABI431A peptide synthesizer produced by PE Company in the United States. The brief description is as follows: standard Fmoc protocol was adopted, arginine was coupled twice, and the peptide chain was extended from the carboxy-terminus to the amino-terminus according to the peptide sequence. After the peptide is synthesized, the corresponding cleavage enzyme is selected for cleavage, and the crude peptide product obtained by removing various protective groups at the same time is stored at -20°C for future use.
[0066] (2) Purification and molecular weight analysis of polypeptide epitopes
[0067] The polypeptide epitopes obtained in the above examples were purified and analyzed by RP-HPLC: each peptide was dissolved in DMSO at a concentration of 20 mg / mL, filtered through a 0.22 μm fiber membrane, purified and ana...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com