Novel regulation protein
A benzene ring-type, polynucleotide technology, applied in the direction of anti-plant immunoglobulin, plant phenotype improvement, introduction of foreign genetic material using vectors, etc., can solve the problem of not identifying regulatory genes and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1: Identification and Expression of Transcription Factors Involved in the Regulation of Petunia Flower Odor
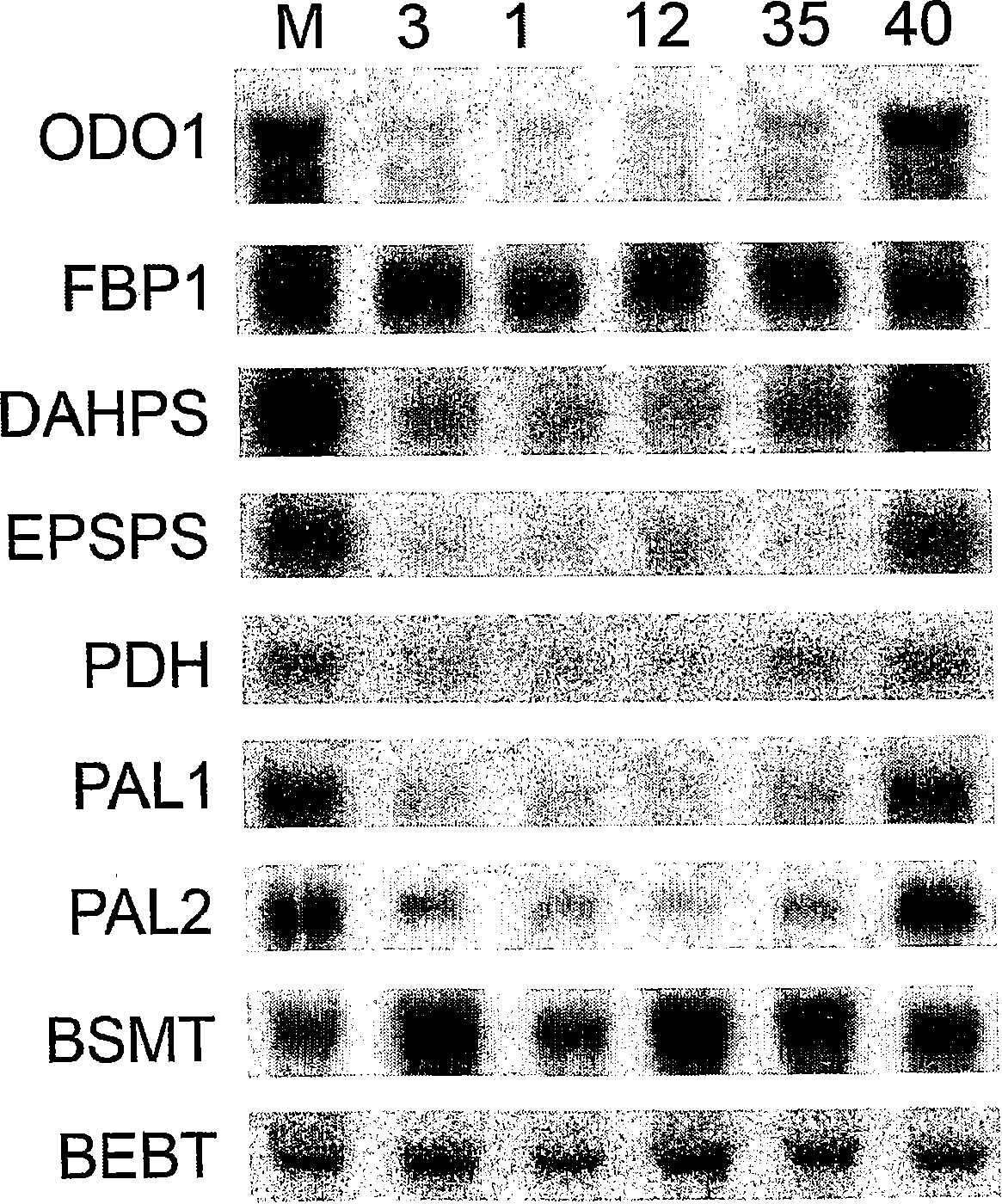
[0067] To identify elements involved in floral odor regulation in petunia, a directed transcriptomics approach was used. Using specialized, highly specific microarrays, the transcriptomes of odorous flowers were compared to those of soon-to-be-scented flowers and non-scented petunia cultivar flowers. Transcription factors were selected for transcription factors that were elevated immediately before odor production and were very low in odorless petunias. One transcription factor, ODO1 (ODORANT 1), is described in detail herein.
[0068] The transcript level of ODO1 increased when volatile benzene ring compounds started to be emitted between noon and 14.00h, which was consistent with its role in regulating flower odor. Transcript levels of ODO1 rose briefly, returning to their lowest levels early the next morning. ODO1 expression is restricted to flower...
Embodiment 2
[0069] Example 2: Characterization of transcription factors involved in regulation of flower odor
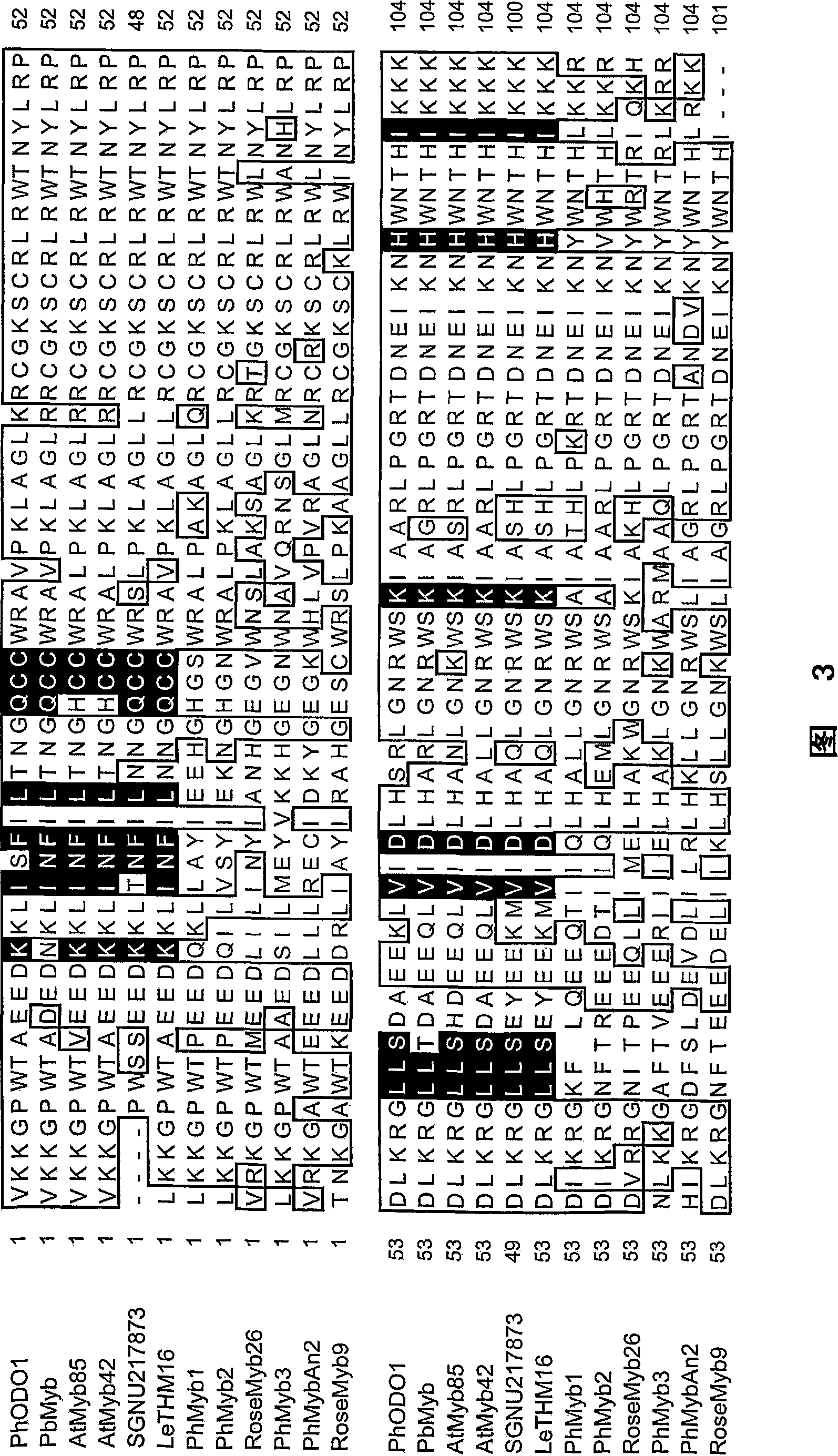
[0070] Sequencing of ODO1 showed that it encodes a 294-amino acid putative protein (SEQID NO.1), which has high homology with R2R3-type MYB family members and has no nuclear localization signal. Although the N-terminal R2R3 domain (amino acids 1-128 of SEQ ID NO.1) contains highly conserved motifs and contains presumably involved in DNA binding to certain variable coremotives (variable coremotives), forming helices- The amino acid of the turn-helix structure, but the C-terminus has no homologous sequence in the Genbank database. Phylogenetic dendrogram analysis showed that ODO1 was most closely related to MYBs from Pimpinella brachicarpa and two MYBs from Arabidopsis thaliana, AtMYB42 and AtMYB85, but the functions of these MYBs were unknown. Seventeen more variable amino acids in the R2R3 domain are conserved among the three proteins.
Embodiment 3
[0071] Example 3: Silencing the ODO1 gene to demonstrate its role in the regulation of flower odor
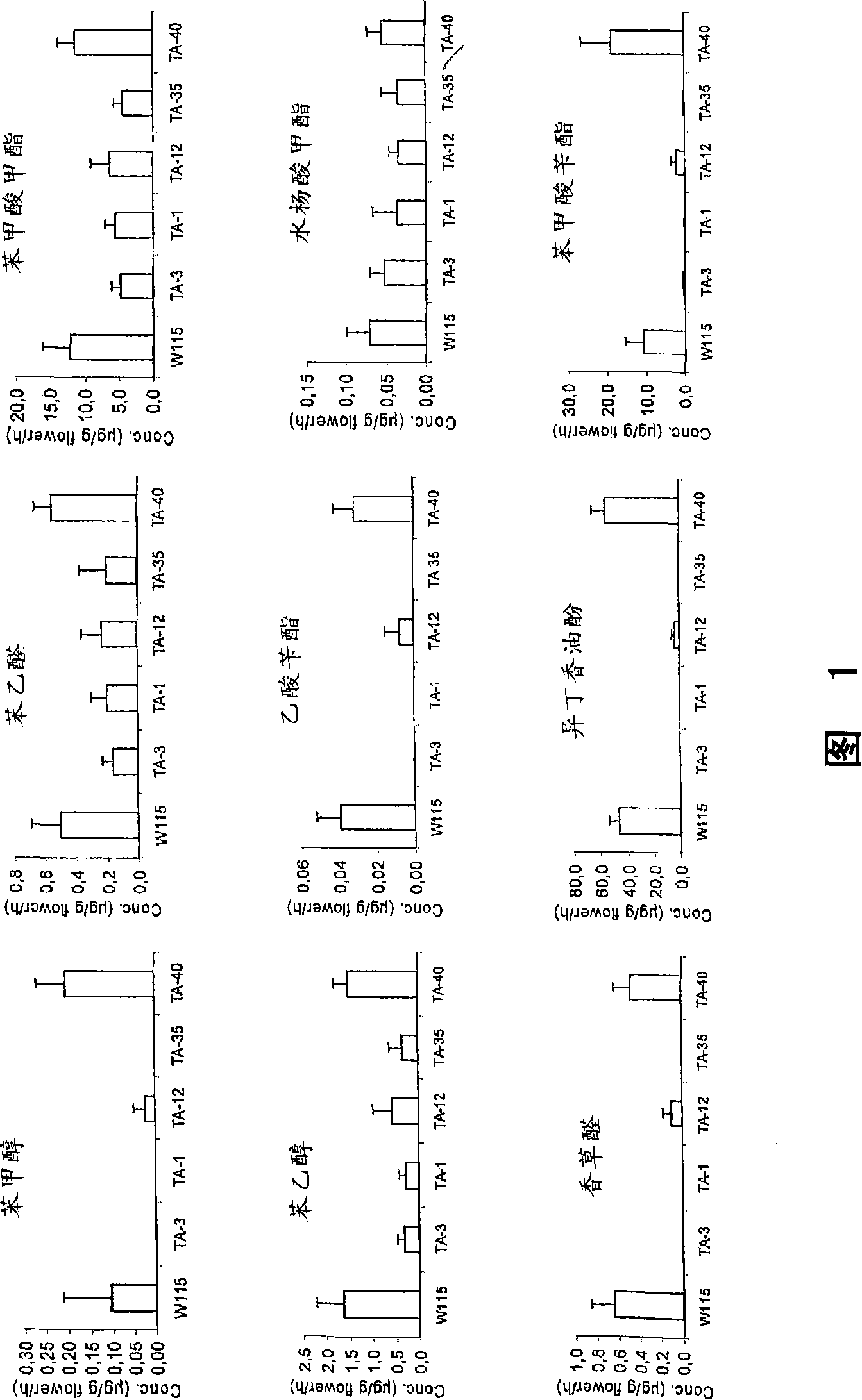
[0072] To investigate the role of ODO1 in the regulation of flower odor, a transgenic approach was employed. Expression of ODO1 in Mitchell was suppressed by RNAi. Since ODO1 is only expressed in flower tubes and petals and not in any other tissue, we used a constitutive promoter in the RNAi construct. This promoter drives the sequence encoding the C-terminus of ODO1 (which has no homology to other genes in the database), and thus is able to inhibit the accumulation of ODO1 transcripts. We used the ODO1 intron in the RNAi construct and also to transform the Mitchell line as a negative control. At 17.00h, the transcript level of ODO1 in the flowers of each independent transformant was analyzed (the parental Mitchell line had a higher transcript level at 17.00h). To rapidly study the volatiles produced by individual flowers of each transgenic line, we used a directed metabolom...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com