Method for testing SNP using restriction enzymes double zyme cutting
A technology of restriction endonuclease and restriction enzyme, which is applied in the field of detection of SNP by restriction endonuclease double digestion, can solve the problems of insufficient detection resolution and slow pre-processing speed, so as to improve detection efficiency and save Detection time, effect of high versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
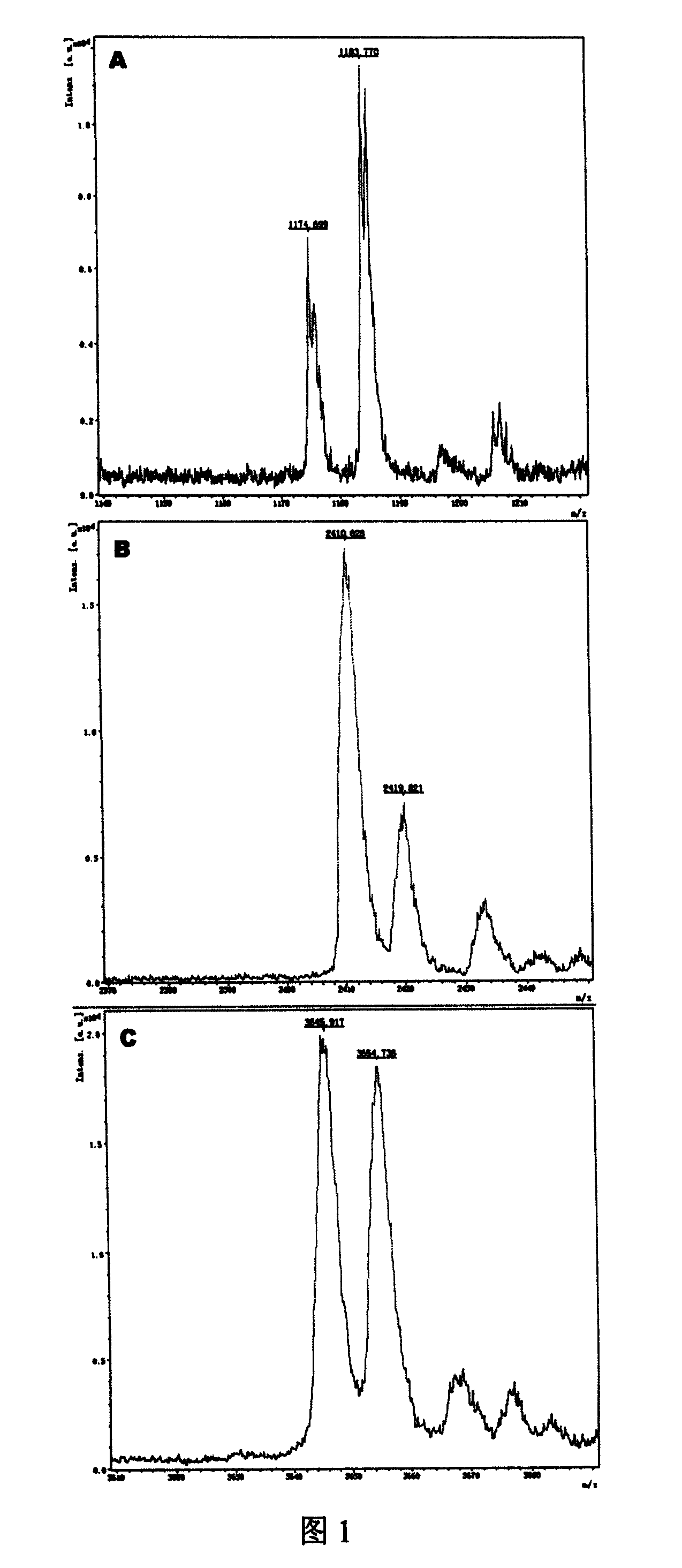
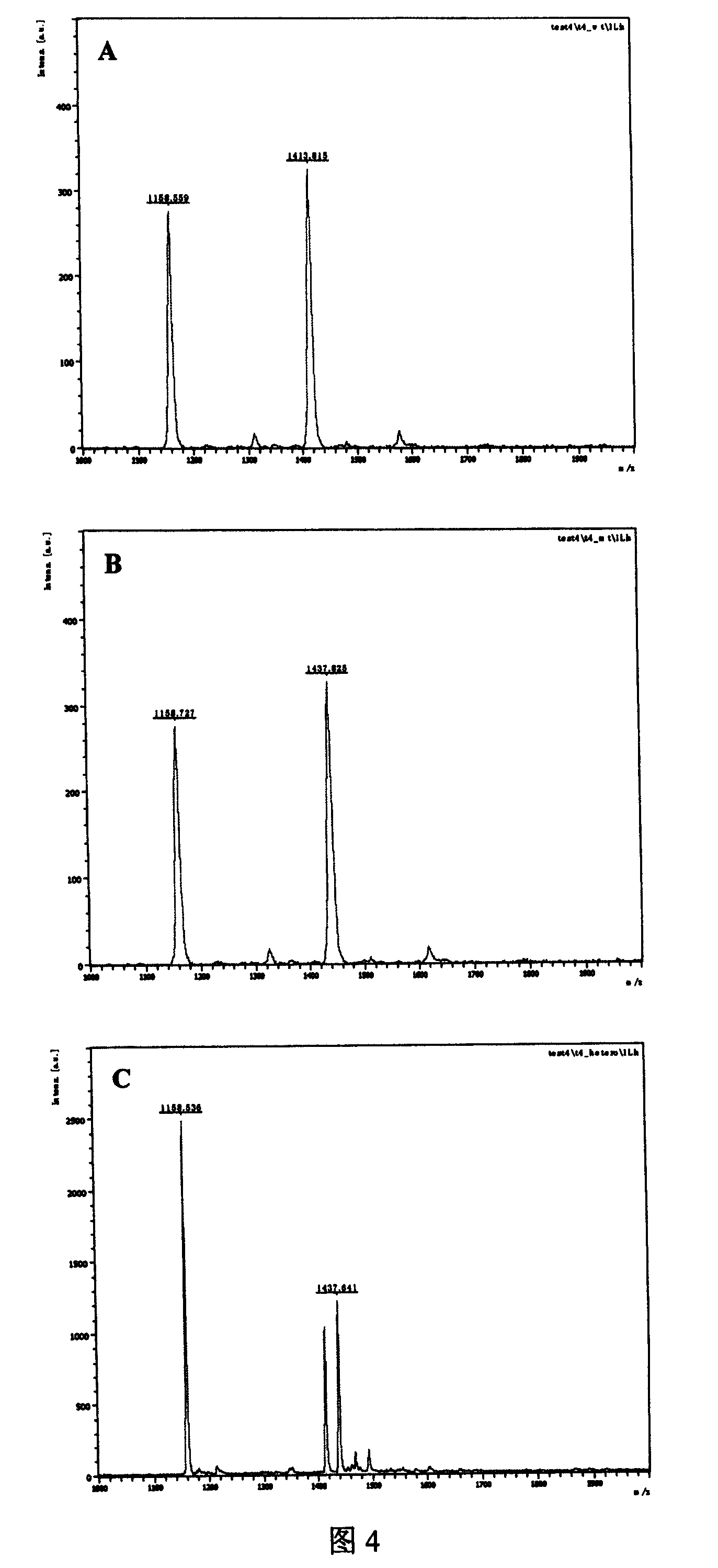
[0045] Embodiment 1: Design and amplify the primer of 669 position SNPs of hepatitis B virus P gene
[0046] Hepatitis B virus (HBV) infection is a serious worldwide health problem, and nearly one million people die from various diseases caused by HBV infection every year. However, for the diagnosed hepatitis B patients, there are multiple SNP mutations in the hepatitis B virus genome in different individuals, which leads to the resistance of patients with the same disease to the same drug. If the SNP markers in hepatitis B patients are studied, the root cause of the differences in the sensitivity of different individuals to the same drug will be revealed, which will help researchers to screen more suitable drugs.
[0047]Among them, the 669th nucleotide of the P gene (NCBI Gene ID: 944565) encoded by the hepatitis B virus genome undergoes a C→A mutation, which changes the encoded amino acid from leucine to methionine (L180M), thereby producing the antiviral drug Lamy Fuding ...
Embodiment 2
[0051] Embodiment 2: Amplify the 669 SNP fragments comprising the P gene of hepatitis B virus
[0052] The size of the PCR reaction fragment is 55 nucleotides, including a 10-nucleotide restriction endonuclease recognition sequence. The PCR reaction system is: 10ul system, including 1.5ul of hepatitis B virus genomic DNA (initial concentration 10ng / ul), 1ul each of forward primer and reverse primer (initial concentration 5pmol / ul), dNTPs 0.8ul (each initial concentration of 2.5mM) , buffer solution 1ul (initial concentration 1x, containing Mg 2+ ), HS Taq enzyme 0.05ul (initial concentration 5U / ul), sterile deionized water 4.65ul; the cycle parameters of the PCR reaction are: 95°C for 5min, then cycle 30 times at 95°C for 20s, 56°C for 20s, and 72°C for 20s , and then 72°C for 7min. After the PCR reaction, the copy number of the nucleotide sequence with the SNP site in the hepatitis B virus genome DNA is amplified. After the completion of the reaction, the PCR product can b...
Embodiment 3
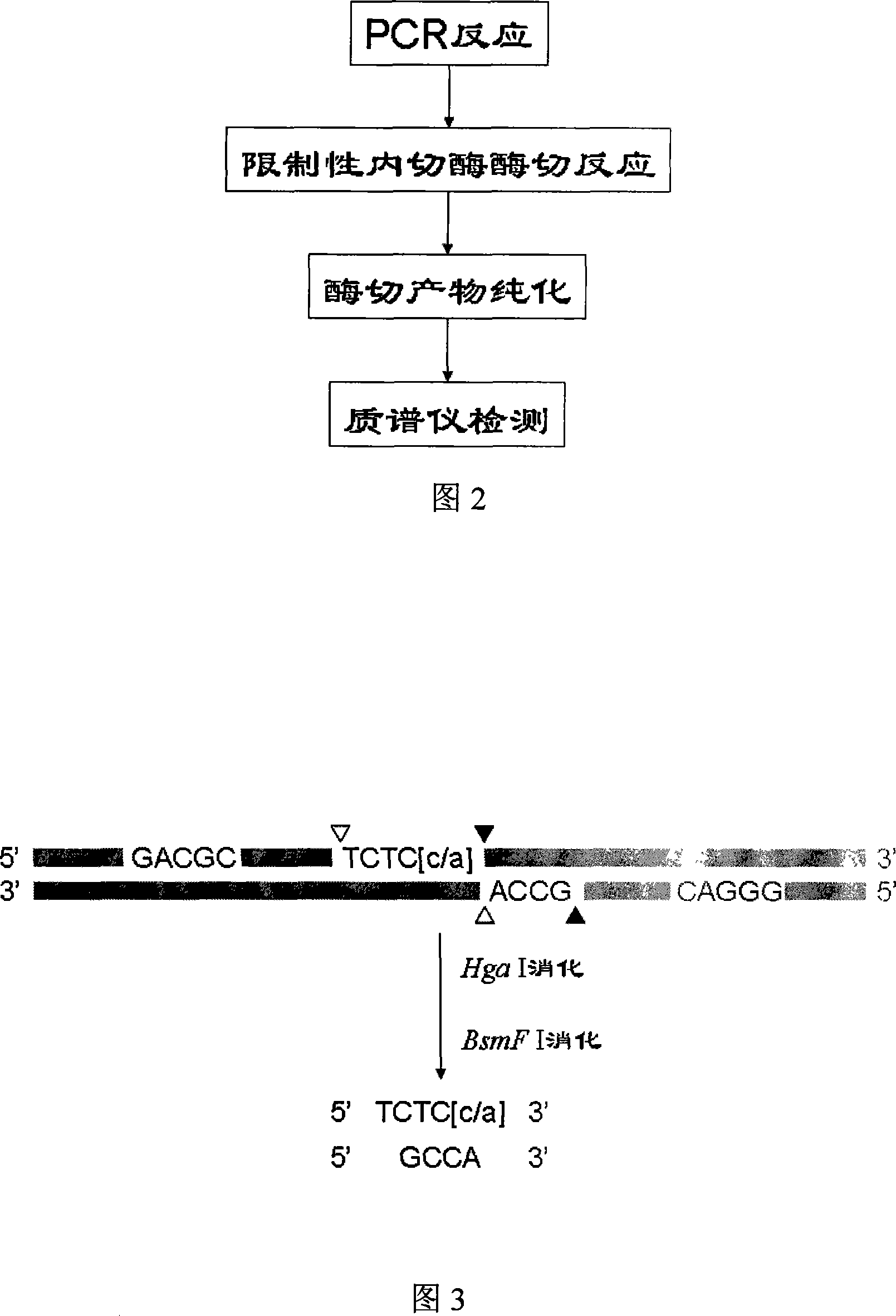
[0053] Example 3: Digestion of amplified products with specific restriction enzymes
[0054] After the PCR reaction, carry out a thorough restriction reaction with corresponding restriction endonucleases. If other restriction endonucleases are used, the reaction system and reaction parameters are slightly different due to the selected restriction endonucleases.
[0055] The first enzyme digestion is a 20ul system, including 10ul of PCR product, 2ul of enzyme buffer (initial concentration 10x), 1ul of HgaI enzyme (initial concentration of 2U / ul), 7ul of sterile deionized water; the enzyme digestion reaction parameters are: 37 ℃ for 40min, 65℃ for 20min; the second digestion is a 30ul system, including 20ul of the first digestion product, 3ul of enzyme buffer (initial concentration 10x), 1.5ul of BsmFI enzyme (initial concentration of 2U / ul), sterile Deionized water 5.5ul; enzyme digestion reaction parameters: 65°C for 40min, 95°C for 20min, ice-water bath for 20min.
[0056] A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com