Method for preparing molecular cloning chip for high-throughput cloning of nucleic acid molecule
A technology for nucleic acid molecules and molecular cloning, which is applied in the field of high-throughput cloning of nucleic acid molecules and high-throughput molecular cloning chips. It can solve the problems of high cost and small amount of templates for four-color fluorescent sequencing, and achieve easy hybridization and extension detection. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
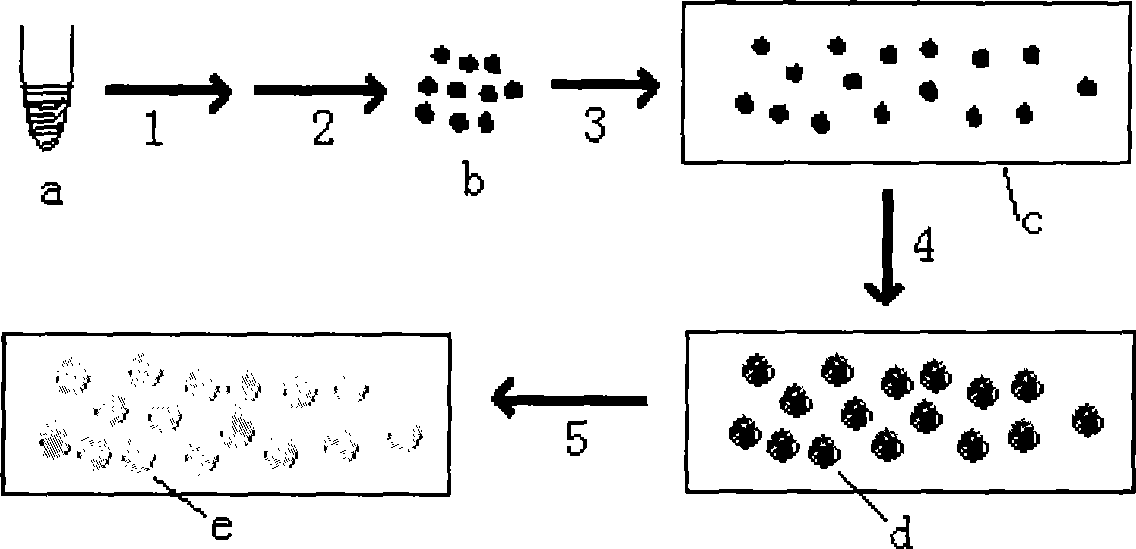
[0025] A method for preparing a molecular cloning chip by high-throughput nucleic acid molecular cloning, the preparation steps are: a. Mixing the nucleic acid molecule template to be cloned with the nucleic acid amplification reaction system reagents to obtain an amplification mixture solution, so that each microliter of the amplification mixture solution The number of nucleic acid template molecules contained is 10 6 ~10 7 , the 5' end of the primer in the nucleic acid amplification reaction system reagent is modified with biotin, aldehyde group, acrylamide group or amino chemical group; b. adding a polymer compound to the above-mentioned amplification mixture solution, the polymer compound The compound can solidify the solution into a gel after lowering the temperature or heat treatment; c. adding twice its volume of the oil phase to the solution obtained in step b for emulsification to obtain a water-in-oil emulsion, the particle size of the emulsion particles 5 μm ± 2.5 ...
Embodiment 2
[0026] Example 2 Preparation of Nucleic Acid Molecular Cloning Array Using Emulsion PCR Amplification and Agarose Gel Particle Fixation
Embodiment approach
[0027] Concrete implementation scheme is as follows:
[0028] Preparation of genomic amplification templates with linkers on both sides:
[0029] Ultrasonic treatment of genomic DNA: choose an ultrasonic instrument with a power of 1200w, and randomly break the genomic DNA into gene fragments of 200bp-600bp size.
[0030] Both ends of genomic DNA fragments are connected with universal linkers: the random fragments obtained by ultrasound are purified to remove some small and broken fragments, and then T4 polynucleotide kinase, T4 DNA polymerase, and Klenow large fragment DNA polymerase are used respectively I and Taq polymerase treatment to make random fragments generate a TA sticky end, and then connect with a universal linker (linker1: 5'-GTCGGAGGCCAAGGCGGCCGT3'linker2: 5'-CGCCTTGGCCTCCGACT-3'), so that the two ends of the genomic fragments are connected a common linker.
[0031] Amplification of the genome flanked by linkers in emulsion PCR with agarose and purification of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com