Method for detecting small molecule RNA chip by ELISA
An enzyme-linked immunosorbent assay and chip technology, which can be used in biochemical equipment and methods, measurement devices, analytical materials, etc., and can solve problems such as limiting the wide application of miRNA chips.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
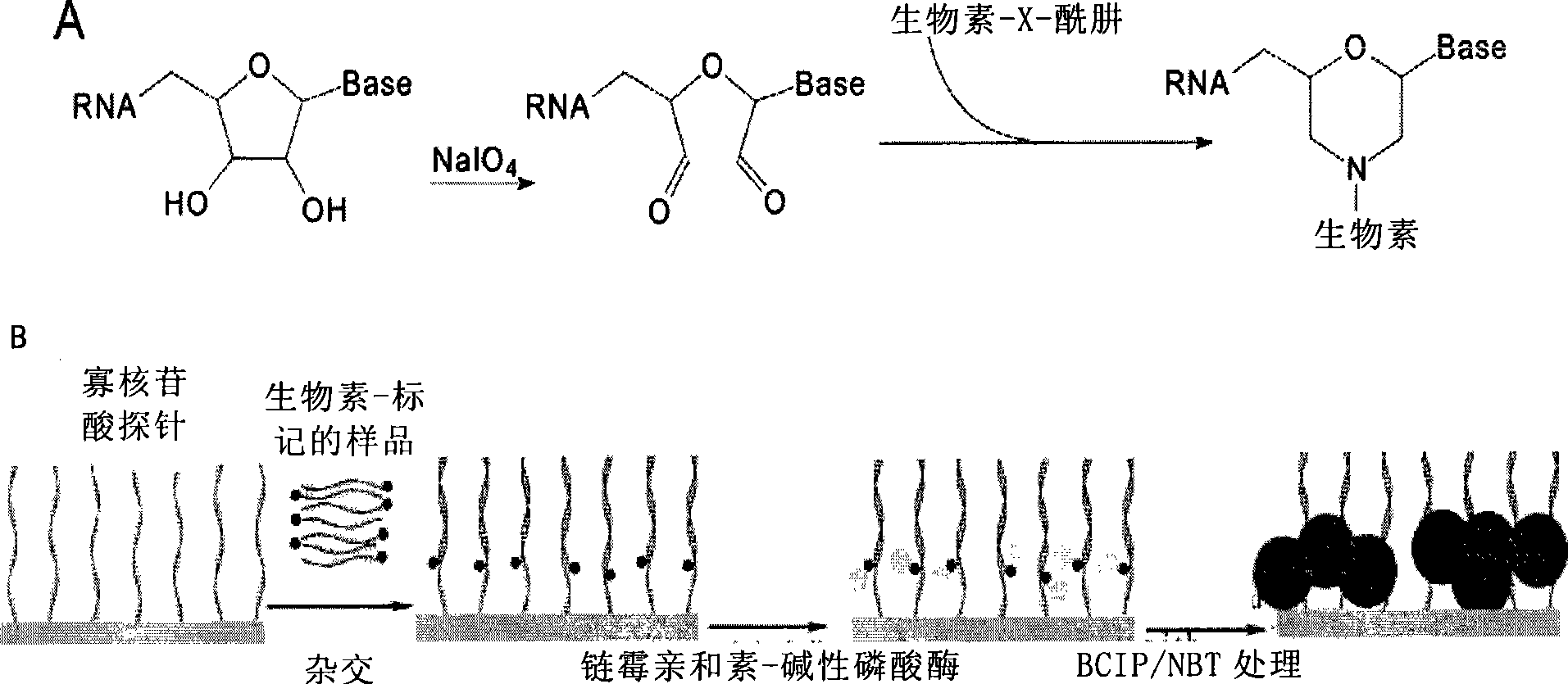
[0055] The preparation method of miRNA chip is known in the art, and generally, miRNA chip comprises conventional solid-phase support (being substrate), and the oligonucleotide probe that is positioned at sample area on substrate, and described probe can pass through for example Covalently immobilized on the solid support. The spotting probe may be an oligonucleotide sequence complementary to any kind of miRNA or an oligonucleotide sequence corresponding to the miRNA sequence. As a preferred mode of the present invention, the probes on the miRNA chip contain sequentially from 5' to 3': amino-(A) n - a nucleotide sequence complementary to miRNA; wherein n=5-20 (preferably 8-12, eg 10). Described solid phase carrier is exactly the substrate material that has oligonucleotide probe on it, and common carrier includes: glass flake, plastic flake, nitrocellulose (NC) membrane, PDVF membrane etc., wherein particularly preferred It's a glass slide.
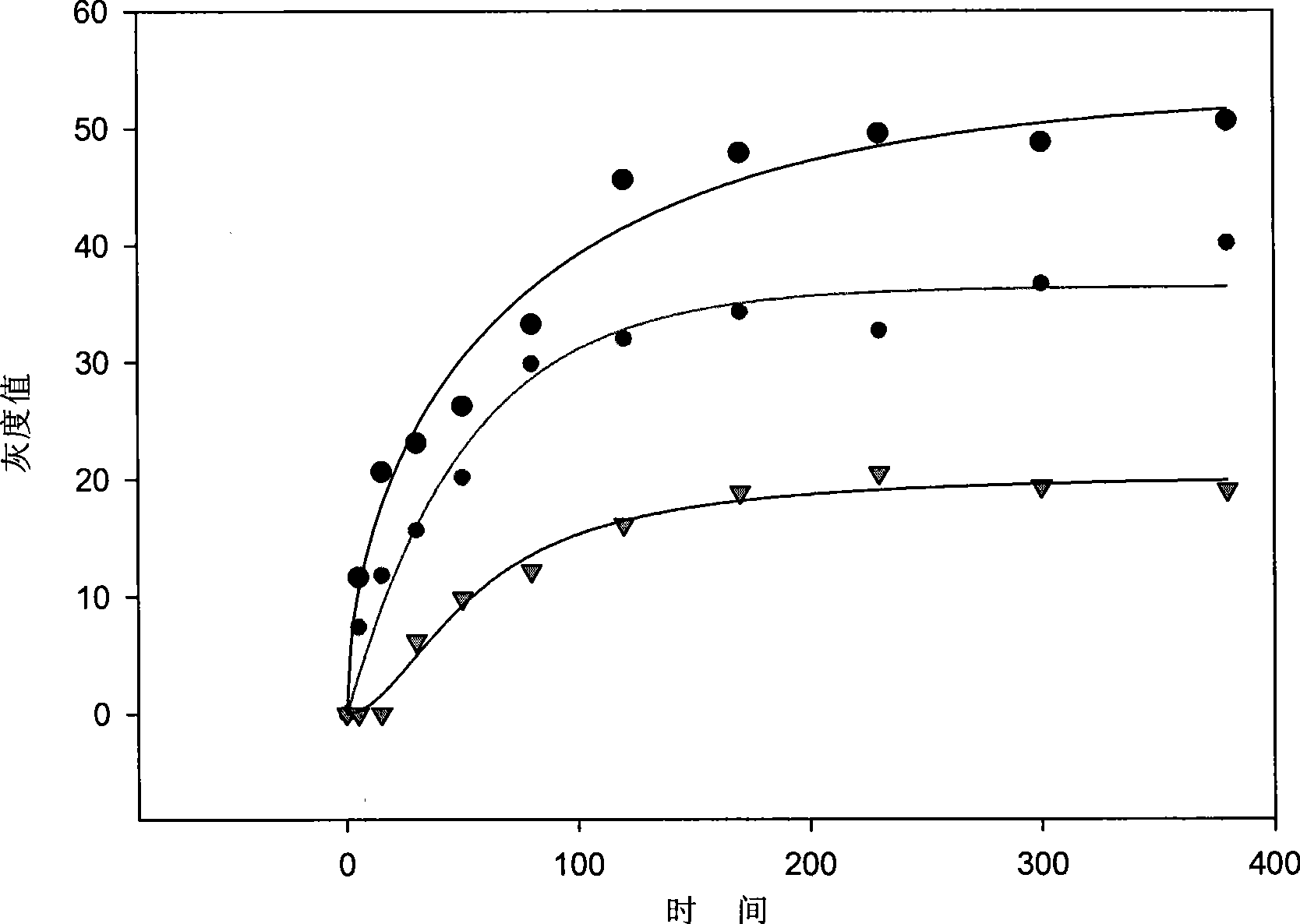
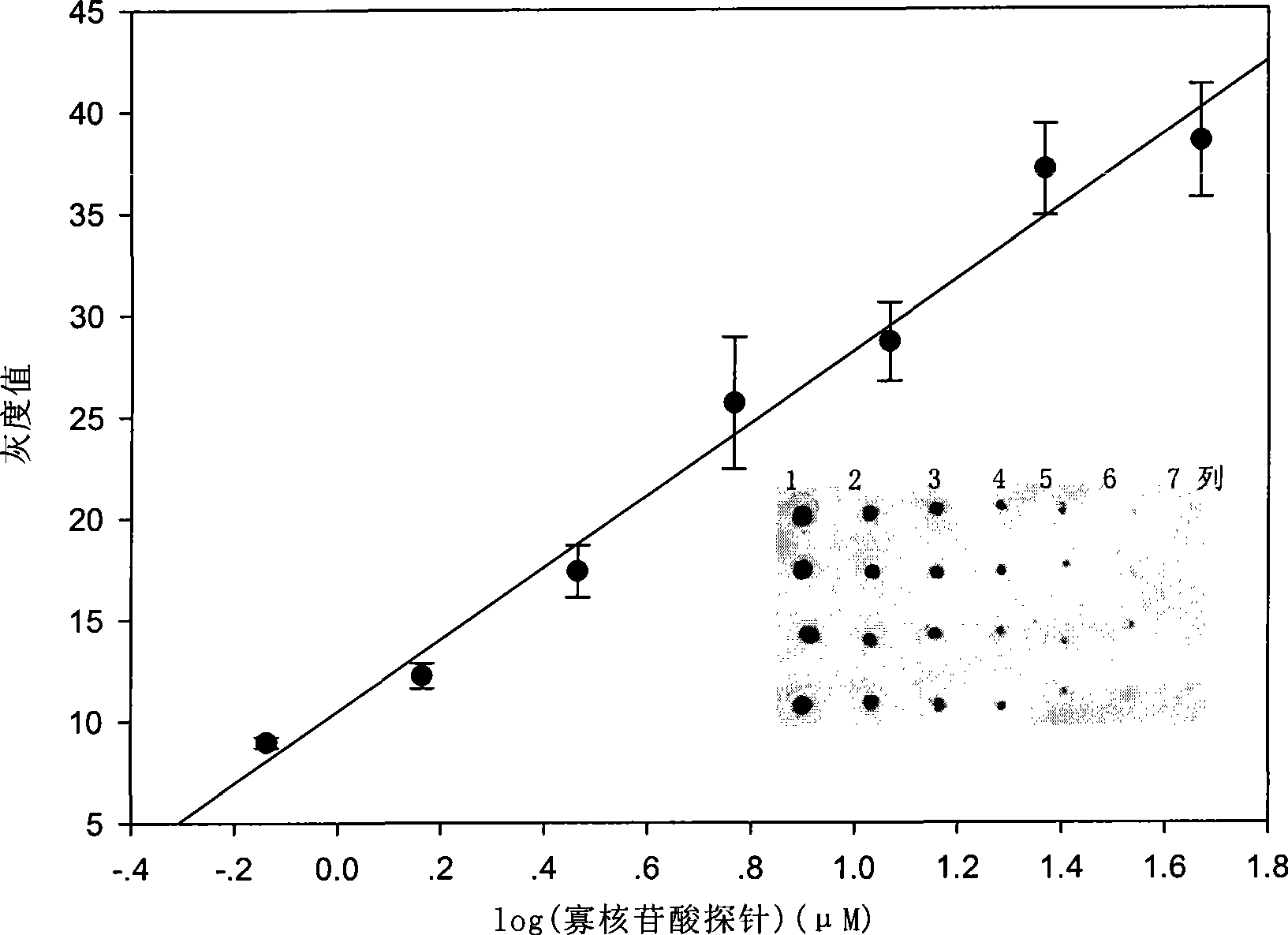
[0056] Based on the ELISA techno...
Embodiment 1
[0070]Example 1 Preparation of rat miRNA chip
[0071] Material
[0072] Glass slides and (3-glycidyloxypropyl)-trimethoxysilane [(3-glycidyloxypropyl)-trimethoxysilane, GOPTS] were purchased from Sigma. Avidin-coupled alkaline phosphatase was purchased from R&D Company, and BCIP / NBT was purchased from Calbiochem Company. DNA probes were synthesized at Invitrogen. The RNA extraction reagent was RNArose from Huashun Company. The rats used in the experiment were 9-week-old male SD rats purchased from Shanghai Experimental Animal Center.
[0073] equipment
[0074] 0lympus C-4000Z digital camera, ordinary microscope. The biochip spotting instrument is PixSys 5500, a product of Cartesian Company, and the assembly spotting needle is SMP4, a product of TeleChem Company. Laser confocal biochip scanner is Genepix 4000B.
[0075] chip making
[0076] Standard 25mm×75mm glass slides were activated according to the method described in Liang, R.Q. et al., Colorimetric detection of...
Embodiment 2
[0080] Example 2 Obtaining and labeling of rat miRNA
[0081] Use RNArose to extract total RNA from the epididymis of 9-week-old SD rats frozen in liquid nitrogen according to the instructions, electrophoresis on 15% polyacrylamide urea denaturing gel, and recover the polypropylene located between the 18nt and 31nt DNARuler by slicing the gel under ultraviolet light Amide glue. The small fragments of RNA were recovered by dipping, precipitated with ethanol, and stored at -20°C. The biotinylation of small RNA was carried out according to the method described in Liang, R.Q. et al., An oligonucleotide microarray formicroRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res, 2005(33), e17: from 90 μg total RNA The obtained 18 μl of 0.5 μg / μl small molecule RNA was diluted with 9 μl of 0.25M pH 5.6 sodium acetate buffer and 4 μl of DEPC water, added 5 μl of 5 mM sodium periodate solution, and reacted at 25°C for 90 minutes in the dark...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com