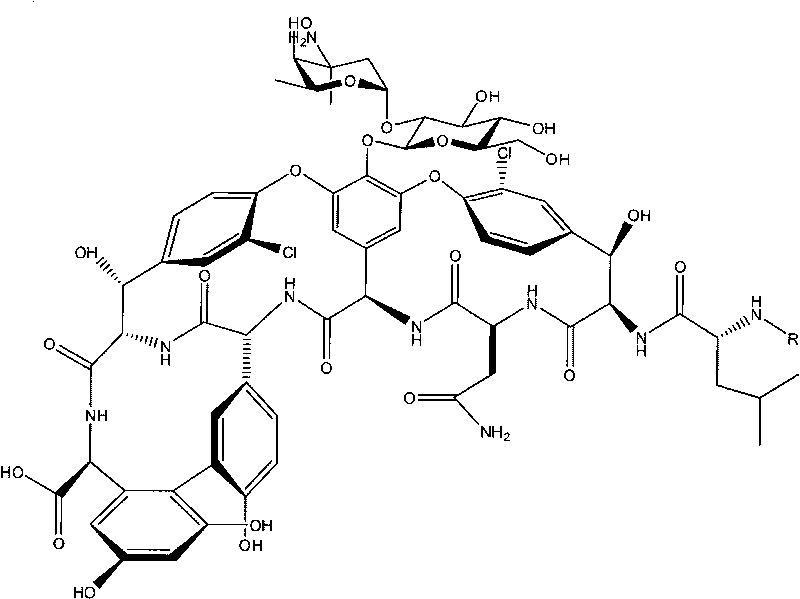
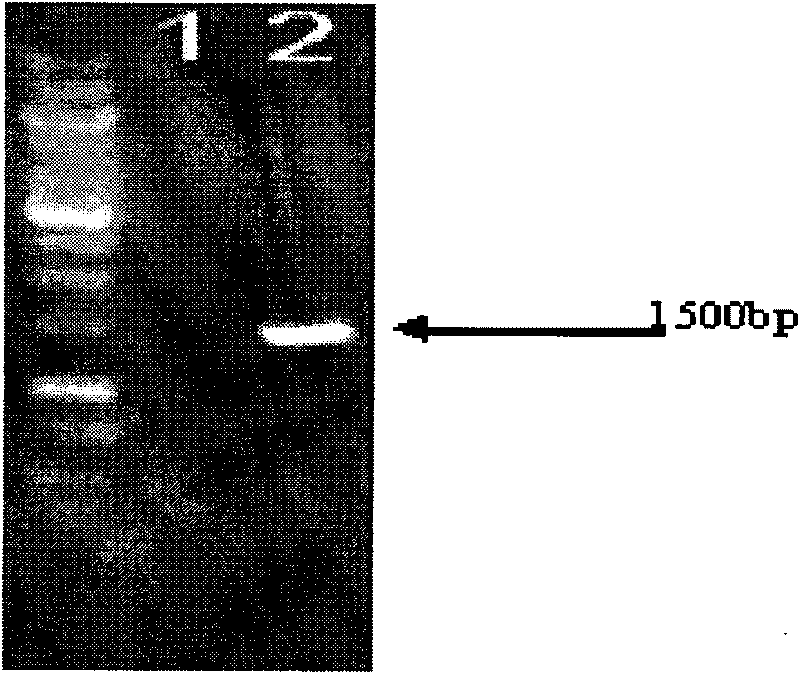
Method for producing norvancomycin
A technology of norvancomycin and vancomycin, applied in the field of genetic engineering, can solve problems such as indeterminate and indeterminate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 , Construction of recombinant plasmid pLY01D
[0042] 1.1. Design and synthesis of primers
[0043] According to the gene sequence of Amycolatopsis orientalis ATCC 43491, two pairs of primers, PNA1 and PNA2 and PNB1 and PNB2, were designed to amplify the fragments of the left homology arm and the right homology arm, respectively. According to the sequence of pHP45omega-aac, the primer pair PNC1 and PNC2 were designed for the amplification of the apramycin (Apr) resistance gene fragment, and the above sequence and its restriction site are as follows:
[0044] PNA1: TCTAGA AAGATCAAGGAAGACCTG;
[0045] wxya
[0046] PNA2: CTGCAG CAGGACAACCTCCAGGTC;
[0047] PstI
[0048] PNB1: CTGCAG TGAGCGCGGCGAGCCTGC;
[0049] PstI
[0050] PNB2: AAGCTT CATCATCGCCGACATCATCGCC;
[0051] Hind III
[0052] PNC1: CTGCAG GGAACTTATGAGCTCAGCCAATCGACT;
[0053] PstI
[0054] PNC2: CTGCAG CTGACGCCGTTGGATACACCA.
[005...
Embodiment 2
[0068] Example 2 , blocking the acquisition of mutant Amycolatopsis orientalis dNMT
[0069] 2.1. Preparation of protoplasts from Amycolatopsis orientalis ATCC 43491
[0070] The mycelia liquid of Amycolatopsis orientalis ATCC 43491 was inoculated into 20 ml of TSB liquid medium, and glycine was added to a final concentration of 2%. Then shake culture at 28° C. for 36-60 hrs, collect the bacterial cells by centrifugation, and wash with Buffer I buffer solution.
[0071] Suspend the mycelium with 10ml of lysozyme solution (1mg / ml, prepared with Buffer I buffer) after filter sterilization, keep it at 28°C for 15 minutes, and then use a sterile funnel equipped with degreasing cotton to remove it by continuous filtration twice. Mycelium. The precipitate was collected by centrifugation, washed twice with Buffer I buffer, and resuspended in 1 ml Buffer I buffer to obtain protoplasts of Amycolatopsis orientalis ATCC 43491.
[0072] 2.2. Preparation of DNA for protoplast transfor...
Embodiment 3
[0087] Example 3 , Amycolatopsis orientalis dNMT strain fermentation culture
[0088] Amycolatopsis orientalis dNMT and Amycolatopsis orientalis dNMT were separated naturally to obtain a single colony.
[0089] Pick a single colony and smear it on the Gaoshi No. 1 slant medium, and cultivate it at 28°C for 5 days. 48h. Then, under aseptic conditions, transfer the cultured seed solution into a fermentation shaker flask with an inoculum amount of 8%, and vibrate and culture at 28°C and 220rpm for 115-120h.
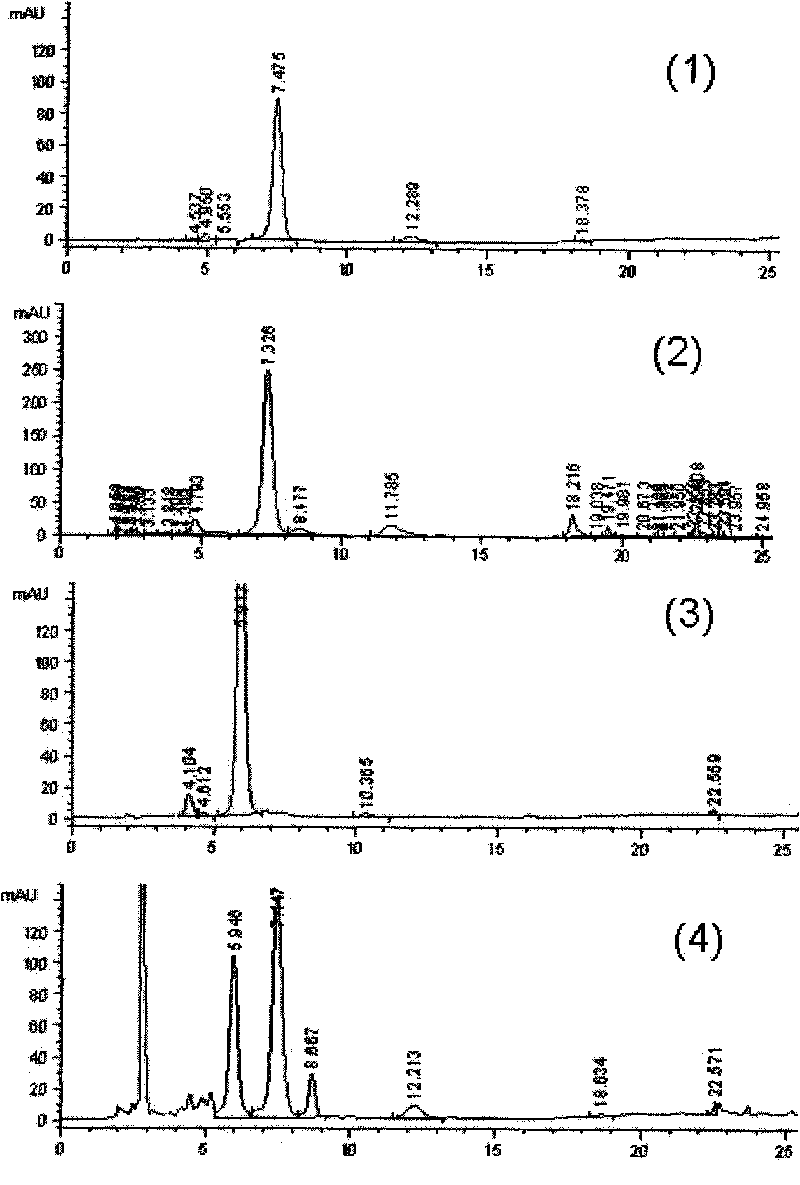
[0090] Get the fermentation broth, centrifuge at 12000rpm for 10min, take the supernatant, and carry out HPLC detection, the detection conditions are as follows:
[0091] Mobile phase preparation: use 0.2% triethylamine solution and phosphoric acid to adjust the pH to 3.2 as a buffer. The buffer solution is mixed with acetonitrile and tetrahydrofuran in a ratio of 92:7:1 and shaken as solution A; the buffer solution is mixed with acetonitrile and tetrahydrofuran in a ra...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com