Detection reagent kit and detection method of soybean phytophthora
A technology of Phytophthora sojae and a kit, which is applied in the biological field, can solve the problems of long cycle time and poor specificity, and achieve the effects of high accuracy, high sensitivity, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1: Design, synthesize primer and establish the PCR reaction system of soybean Phytophthora detection kit
[0030] 1. Primer design and synthesis
[0031] Design of PsYpt3F / PsYpt2R specific primers for Phytophthora sojae:
[0032] PsYpt3F (20bp): 5'-TACCAATAATCAGAAGCGTA-3' (SEQ ID NO.1);
[0033] PsYpt2R (18bp): 5'-CCTTGTCTGCCCTCTCGA-3' (SEQ ID NO. 2).
[0034] At the same time, the Ypt1 universal primer Yph1F / Yph1R of Phytophthora (see the nucleotide sequence table SEQ ID NO.3 and SEQ ID NO.4 for details) was also designed as a round reaction primer for nested PCR:
[0035] Yph1F (20bp): 5'-CGACCATTGGCGTGGACTTT-3' (SEQ ID NO.3);
[0036] Yph1R (20bp): 5'-ACGTTCTCGCAGGCGTATCT-3' (SEQ ID NO.4).
[0037] All primers were synthesized by Invitrogen Company.
[0038] 2. Establish PCR reaction system
[0039] 1. Establish a conventional PCR reaction system
[0040] 1mL conventional PCR reaction system includes: several templates, 0.05mmol Tris.Cl, 0.125mmol K...
Embodiment 2
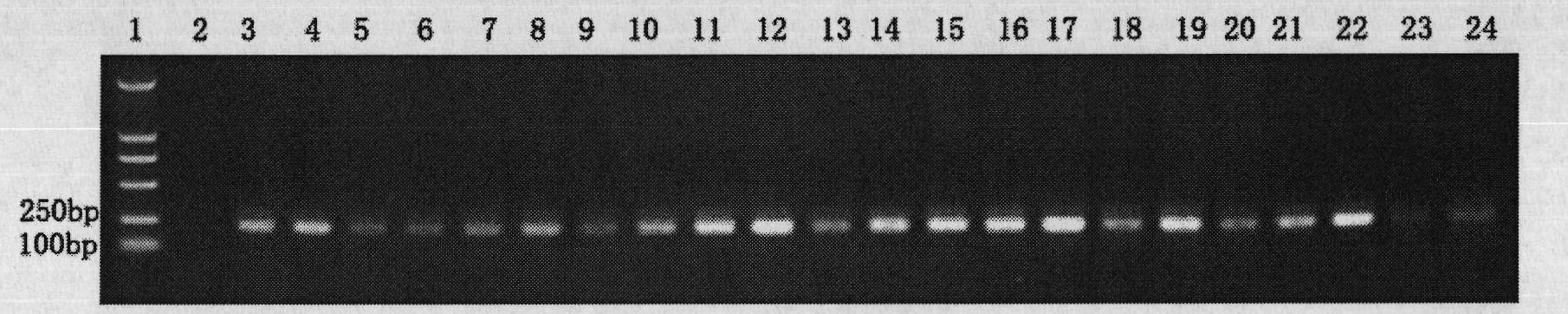
[0045] Example 2: Detection of Phytophthora sojae in soil samples
[0046] Phytophthora sojae was detected from soil samples of imported soybeans with fungus at the customs. The detection process is as follows:
[0047] 1. Enrichment of oospores in soil:
[0048] Take 20-100 grams of the soil sample to be tested, grind it, first use a 200-mesh sieve to remove larger soil particles, then filter through a 400, 500, and 800-mesh sieve, and wash it repeatedly with 3-10 liters of water at the same time. Collect oospores on the net and suspend with 1ml sterile water. Since oospores cannot pass through the 800-mesh sieve, such treatment can achieve the effect of enriching oospores.
[0049] 2. DNA extraction from trace oospores:
[0050] Transfer the oospores suspended in sterile water to a 1.5mL centrifuge tube, at 12000r.min -1 Centrifuge at high speed for 5 minutes and pour off the liquid;
[0051] Add 50 μL CTAB buffer, grind, then add 500 μL CTAB buffer, and bathe in water ...
example 3
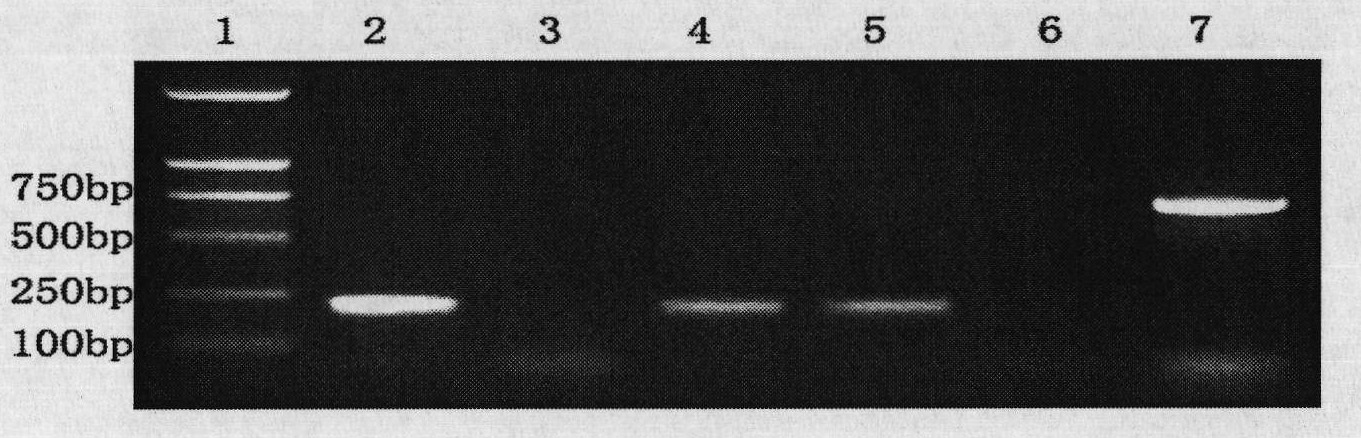
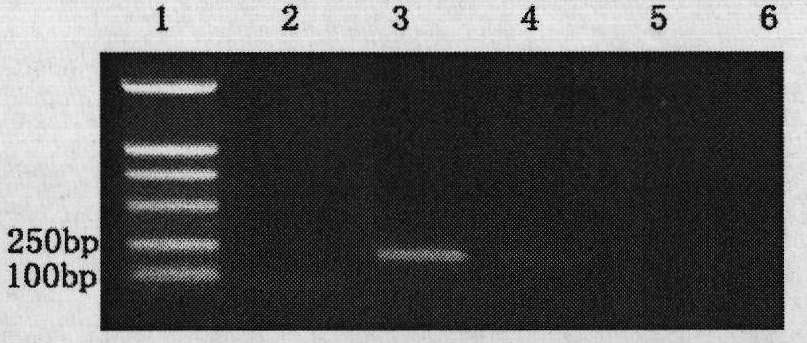
[0064] Example 3: Detection of Zoospores of Phytophthora sojae from Contaminated Water
[0065] Take 500mL of irrigation water contaminated by Phytophthora sojae, centrifuge it for 20min under the centrifugal force of 6000g, discard the supernatant, suspend the precipitated zoospores with 100uL water, transfer them into a 1.5mL centrifuge tube, add 0.05g of quartz sand, and vortex 10sec, take 1uL zoospore fragmentation liquid as template, carry out gene amplification according to the method for embodiment 2, can amplify the unique fragment of Phytophthora sojae, the results are shown in Figure 3a and Figure 3b . in, Figure 3a Sensitivity of primers PsYpt3F / PsYpt2R to amplify different concentrations of zoospores; Figure 3b The sensitivity of the nested PCR reaction to amplify different concentrations of zoospores for the combination of primers Yph1F / Yph2R and PsYpt3F / PsYpt2R; lane 1 is a 2000bp DNA marker; lane 2 is a negative control; lanes 3-6 are 25μl reaction system...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com