Preparation method of membrane protein polypeptides for improving detection rate of membrane proteins
A technology of membrane protein and detection rate, applied in the field of preparation in the field of biological detection technology, achieves the effects of low cost, short time consumption and simplified preparation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
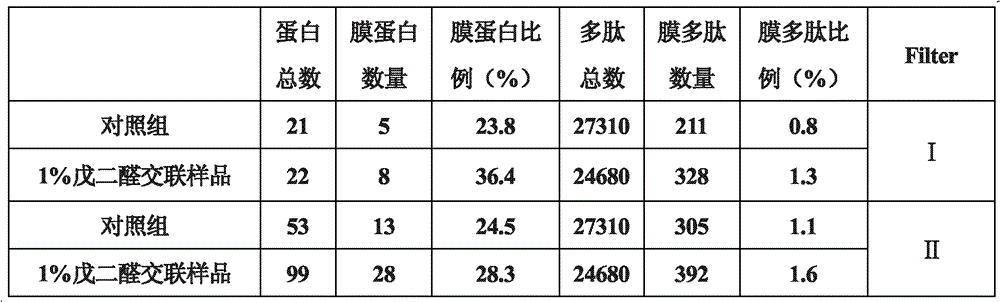
[0023] Kunming rats (male, weighing 250-300 grams, purchased from Shanghai Slack Experimental Animal Center) took blood from the orbit, put it into a test tube previously filled with 2% potassium oxalate solution, and mixed quickly. Centrifuge at 1000g at 4°C for 5 minutes, and use a pipette to remove the supernatant and the white blood cell film on the upper layer of red blood cells. Slowly mix the precipitated erythrocytes with twice the volume of normal saline, centrifuge again at 1000 g at 4°C for 5 min, discard the supernatant, add normal saline and mix well, and repeat centrifugation for 4 to 5 times. After the last centrifugation of the red blood cells, discard the supernatant, which is the washed red blood cells. Draw 50 μl of red blood cells, add 2450 μl of pre-cooled 1% glutaraldehyde solution (the final concentration of red blood cells is 2%) in an ice bath, and slowly rotate and react at 4°C for 0.5 hours. After the reaction, centrifuge at 1000 g for 5 min at 4°C....
Embodiment 2
[0027] Take 50 μl of washed mouse red blood cells, add 2450 μl of pre-cooled 0.1% glutaraldehyde solution (final concentration of red blood cells is 2%) in an ice bath, and slowly rotate and react at 37° C. for 0.5 hours. After the reaction, centrifuge at 1000 g for 5 min at 4°C. The precipitated cells were washed 3 times with normal saline in the same way. Erythrosomes cross-linked with 0.1% glutaraldehyde were obtained. Take 10 μl each of untreated erythrocytes, 1% glutaraldehyde cross-linked and solidified erythrocytes prepared in Example 1, and 0.1% glutaraldehyde cross-linked erythrocytes (about 8.7×10 7 ), add 490 μl 10 mmol PBS (pH7.4), add 10 μl 0.1 μg / μl sequencing grade trypsin. Enzymatic hydrolysis in a 37°C oven for 14-16 hours. Collect the enzymatic peptides, use C18Zip-tips to desalt and enrich the peptides. Samples were lyophilized for LC-LTQ analysis. The obtained mass spectrometry data was searched using SEQUEST; Trans-Proteomie Pipeline (ISB / SPC Proteomi...
Embodiment 3
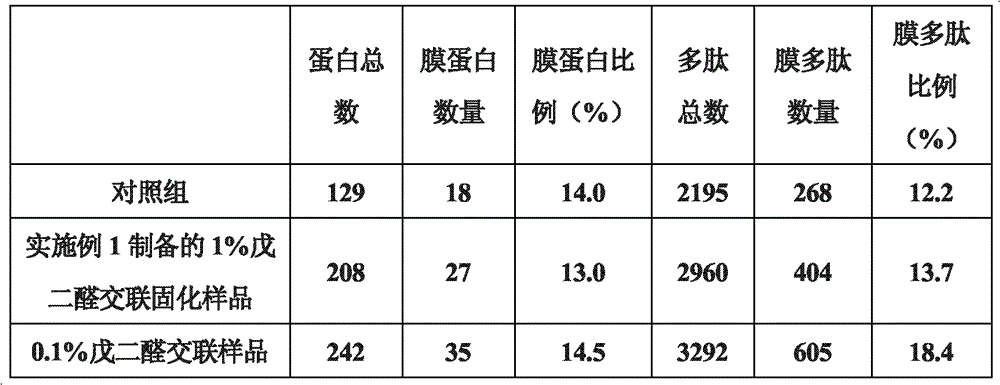
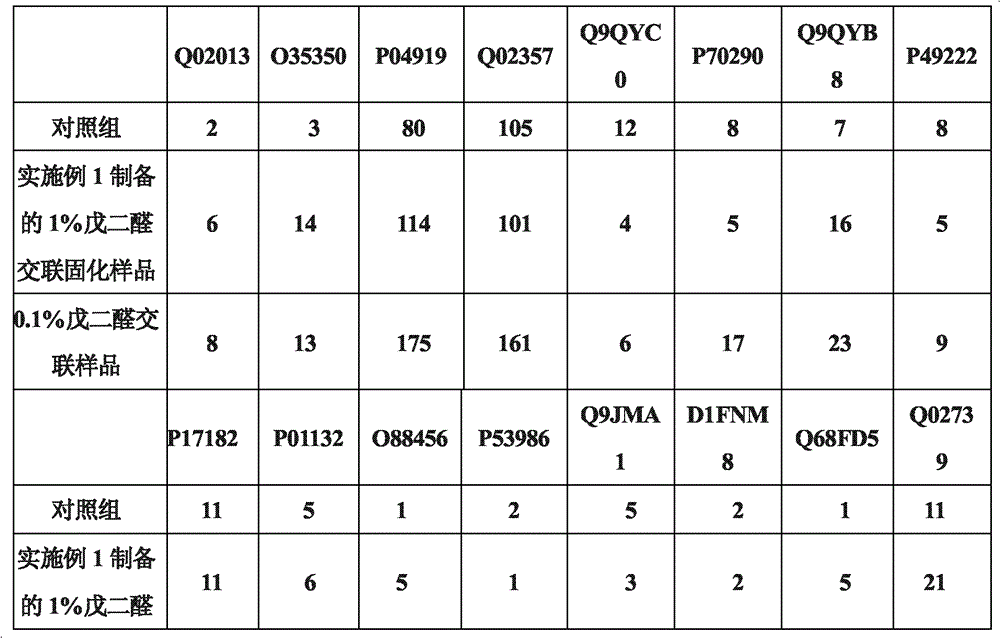
[0036]Get each 10 μl of untreated erythrocytes and 0.1% glutaraldehyde cross-linked and solidified erythrocytes prepared in Example 2 (about 8.7×10 7 ), add 490 μl 50 mmol Tris-HCl buffer, pH 8.0, add 10 μl 0.1 μg / μl sequencing grade endoproteinase GluC (Endoproteinase Glu-C). Enzymatic hydrolysis at 25°C for 14-16 hours. Collect the enzymatic peptides, use C18 Zip-tips to desalt and enrich the peptides. Samples were lyophilized for LC-LTQ analysis. The obtained mass spectrum data was searched using SEQUEST; Filter was set to ΔCn>=0.100; Xcorr(±1, 2, 3)=1.90, 2.20, 3.75; #matches=1. Results A higher proportion of membrane proteins was detected in the sample prepared in Example 2 and treated with 0.1% glutaraldehyde cross-linking and curing. Table 5 shows the number and ratio of membrane polypeptides and membrane proteins detected before and after glutaraldehyde crosslinking:
[0037] table 5
[0038]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com