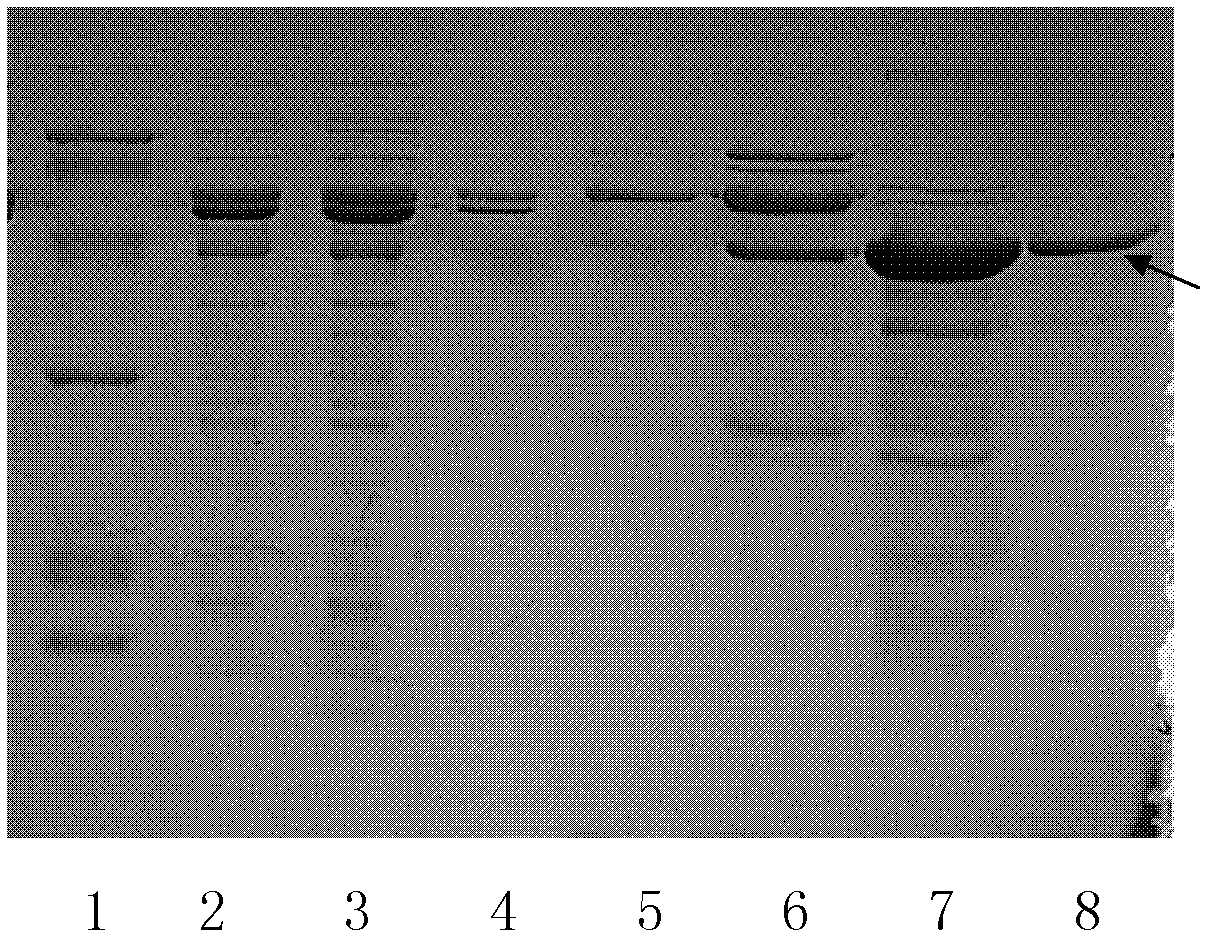Method for preparing enzyme through coexpression of recombinant protease and molecular chaperone
A molecular chaperone and protease technology, applied in the field of recombinant protein expression, can solve problems such as difficult to obtain soluble expression of protease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0028] 1. Gene source
[0029] Protease sequences are available via the Uniprot protein database under accession numbers E4RD31, B7V5W2, Q52078. Perform reverse transcription on the amino acid sequence of the protease to obtain the gene nucleotide (DNA) sequence, and optimize the codon preferred by Escherichia coli for the DNA sequence (codon optimization is completed on the following website: http: / / www.jcat.de / ) . Gene synthesis codon-optimized gene sequence, sequenced correctly, to obtain the target gene. The amino acid sequence whose sequence number is E4RD31 is shown in SEQ ID NO.1, and its nucleotide sequence is shown in SEQ ID NO.2; the amino acid sequence whose sequence number is B7V5W2 is shown in SEQ ID NO.3, and its nucleotide sequence The sequence is shown in SEQ ID NO.4; the amino acid sequence of Q52078 is shown in SEQ ID NO.5, and the nucleotide sequence is shown in SEQ ID NO.6.
[0030] 2. Construction of expression vector pCold recombinant plasmid
[0031]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com