SSR (Simple Sequence Repeat) molecular marking method for identifying variety authenticity and/ or variety purity of high-quality transgenic hybrid cotton CCRI (Chinese cotton research institute) 70
A technology of variety purity and labeling method, which is applied in the field of biotechnology applications, can solve the problems of large environmental impact on morphological identification, heavy identification workload, and seasonal restrictions, and achieve stable and reliable detection results, stable genetics, and simple and convenient operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Methods: Seed DNA was extracted, PCR was amplified using SSR molecular markers, the amplified products were subjected to gel electrophoresis, the results were counted and characteristic primers were screened.
[0027] 1. Extract cotton genomic DNA.
[0028] The experimental material in this example is the high-quality insect-resistant hybrid cotton "Zhongmian 70" commercial seeds (provided by Hunan Longping Hi-Tech Yahua Cotton Oil Seed Industry Co., Ltd.).
[0029] (1) Remove the outer hard shell of a single cotton seed, thoroughly crush it, transfer it to a 2 mL centrifuge tube, add 800 μL of SDS extract (composition: 1% SDS, 0.01mol / L EDTA, 0.7 mol / L NaCl, 0.05 mol / L Tris, 0.5% sorbitol, 1% PVP, 1% β-mercaptoethanol), after fully mixing.
[0030] (2) Put in a water bath at 65°C for 30 minutes, and shake gently every 10 minutes or so.
[0031] (3)) Add an equal volume of 800 μL phenol:chloroform:isoamyl alcohol (25:24:1), mix up and down until there is no layer...
Embodiment 2
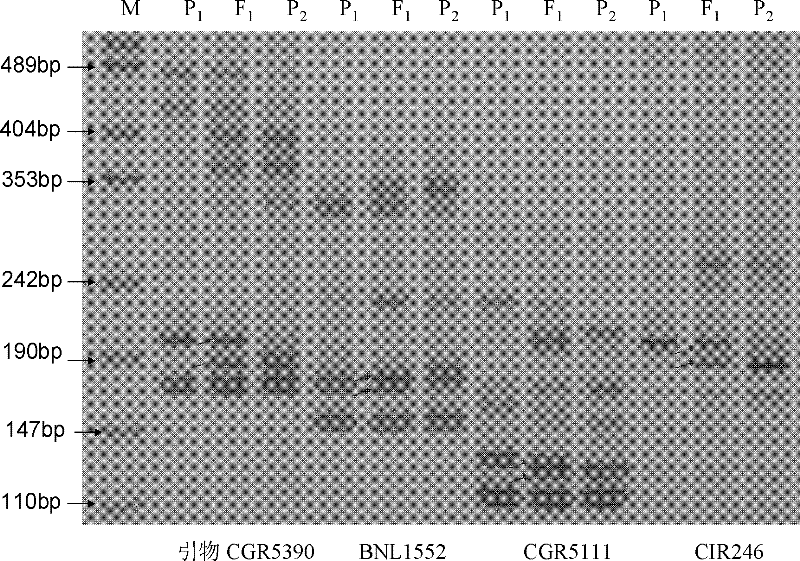
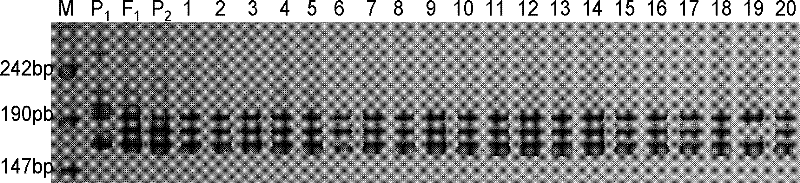
[0054] The four characteristic primers screened out in Example 1 were used to identify the genetic purity of the seeds.
[0055] The experimental material in this example is the high-quality insect-resistant hybrid cotton "Zhongmian 70" commercial seeds (provided by Hunan Longping Hi-Tech Yahua Cotton Oil Seed Industry Co., Ltd.). Generally, 100 seeds are extracted for detection and genetic purity is calculated.
[0056] 1. Cotton genomic DNA was extracted, and the extraction method was the same as in Example 1.
[0057] 2. Use CGR5390, BNL1552, CGR5111 and CIR24 primers, and use the genomic DNA extracted in the first step above as a template for PCR amplification.
[0058] The PCR amplification reaction system is 10 μl, including 6.40 μl ultrapure water, 1.0 μl 10×Buffer (containing Mg 2+ / 20mM), 10mM dNTPs 0.50μ1, 10μM forward primer 0.50μ1, 10μM reverse primer 0.50μ1, 30ng / μ1 template DNA 1.0μ1, 5U / μ1 Taq DNA polymerase 0.10μ1. The PCR reaction program was: pre-denatur...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com