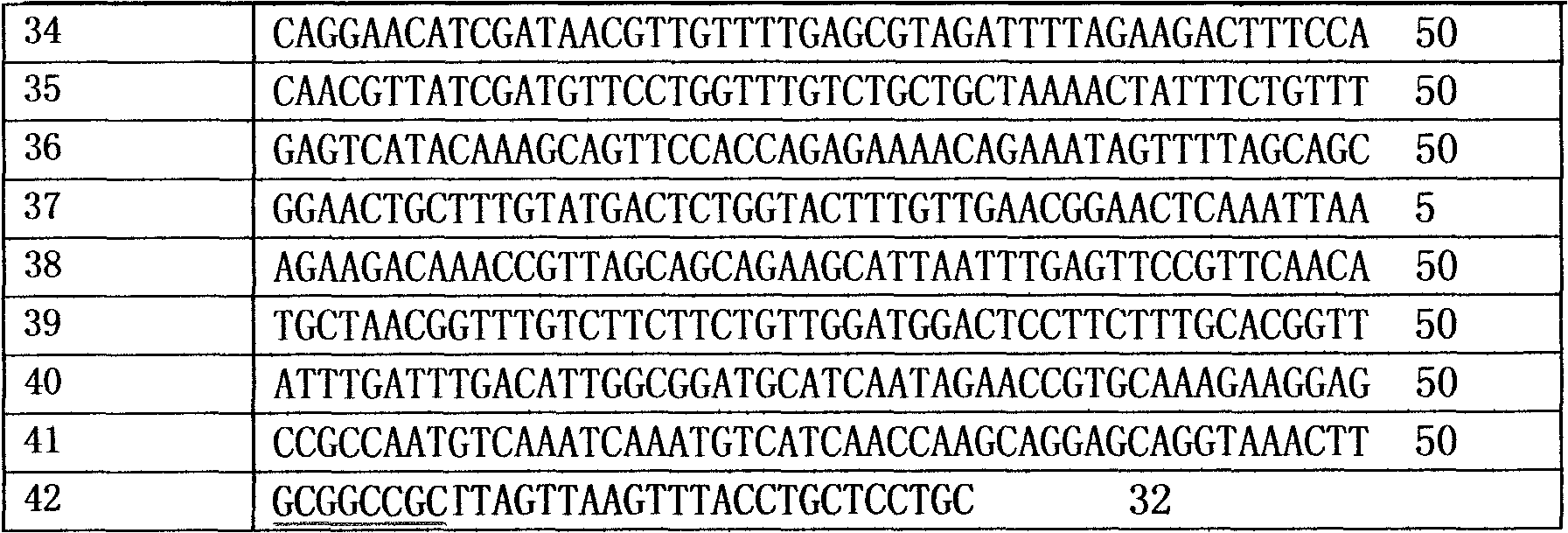Optimized nucleotide sequence of alkaline pectinase pell68s and high-level expression method thereof
A technique for optimizing sequences and pectinase, applied in the biological field, can solve the problem of low production of alkaline pectinase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] The present invention will be further described below in conjunction with specific embodiment:
[0025] The experimental methods used in the following examples are conventional methods unless otherwise specified.
[0026] The materials and reagents used in the following examples can be obtained from commercial sources unless otherwise specified.
[0027] Implementation 1. Synthesis of pel168s optimized gene sequence
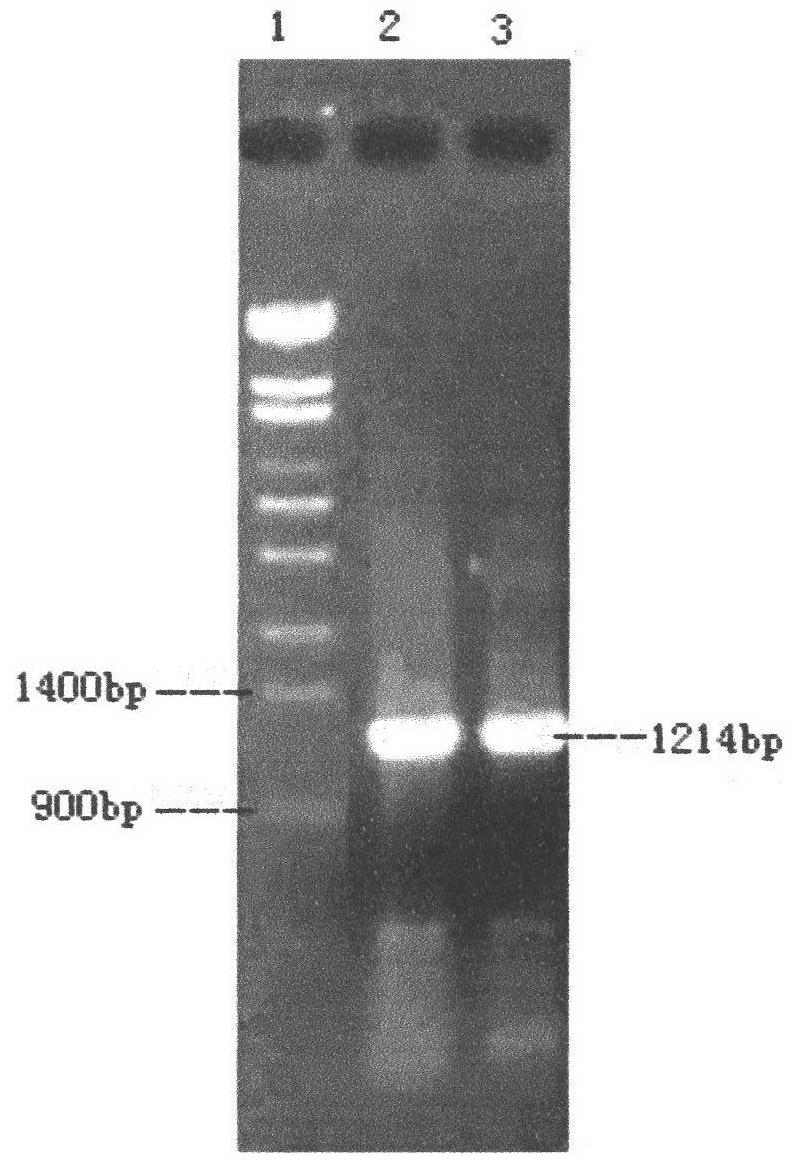
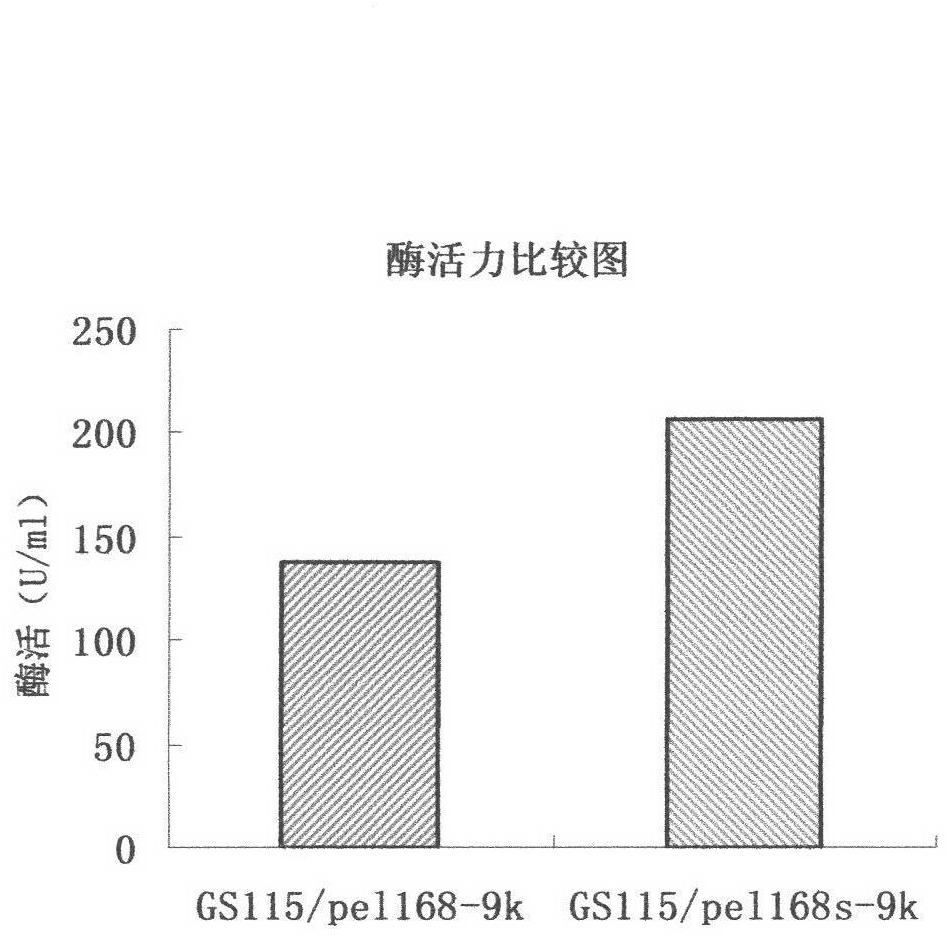
[0028] (1) The routine PCR overlapping primer extension method was adopted in the laboratory. Input the pel168 gene sequence (Bacillus subtilis 168, GenBank accession number: AL009126) into the DNAworks software, select P.pastoris in the codon frequency table (Codon Frequency Table), and select SalI and PmeI in the shielded restriction enzyme site option and other enzyme cutting sites to obtain a series of optimized gene sequences expressed in Pichia pastoris related to pel168s, the best effect is that 294 bases in the sequence are optimized, and the sig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com