Encoding gene of type I pullulanase as well as recombinant expression and application thereof
A kind of pullulanase encoding and pullulanase technology, which is applied to the cloning of the pullulanase encoding gene of Aeromonas sp. In the field of lulanase preparation, it can solve problems such as difficulties in recombinant expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1: obtain by PCR method Aeromonas sp. As pullulanase full-length gene pluAs
[0045] By comparing the reported pullulanase genes of Aeromonas genus (derived from the information obtained after the whole genome sequencing of enzyme-producing strains), look for the conserved regions at the 5' and 3' ends of the gene (see image 3 ), design PCR primers Aero-F and Aero-R as follows:
[0046]Aero-F: ATGGATATGAAAAAGACCAAAGTGGCG (forward primer, without restriction site), that is, SEQ ID NO 3.
[0047] Aero-R: CTTGRCRCKGCACTCCTGRATSACRGCRGTACC (reverse primer, no restriction site), namely SEQ ID NO 4.
[0048] by Aeromonas sp. The As genome was used as a template, and PCR amplification was performed with the above primers. After the PCR amplification product was recovered, it was connected to the pMD18-T vector, transformed into the Escherichia coli Top10 strain, and the plasmid pT-pluAs was obtained, and the sequencing results were performed using the ...
Embodiment 2
[0049] Embodiment 2: obtain by PCR method pluAs- 1 and the preparation of Escherichia coli Rosetta genetically engineered bacteria
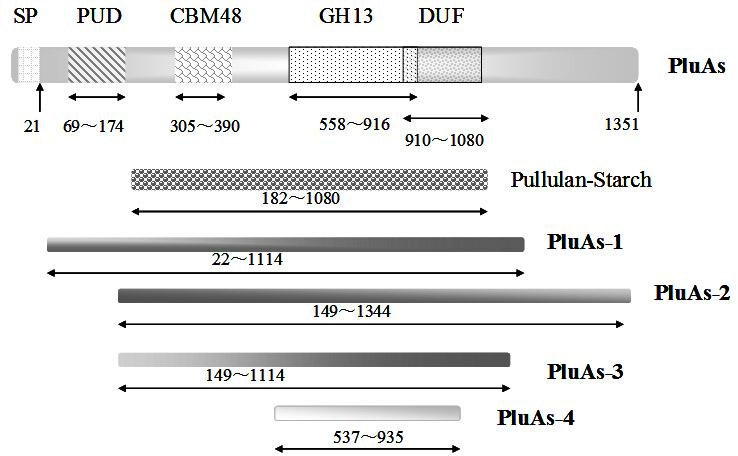
[0050] The present invention has amplified such as by PCR method figure 2 shown pluAs The gene fragments are: the four core domains of the PluAs protein (amino acids 22-1114, PluAs-1), the gene fragments without the PUD domain (amino acids 149-1344, PluAs-2), Gram-negative bacteria The conserved domain of pullulanase (amino acids 149-1114, PluAs-3). The Escherichia coli recombinant expression plasmids are pPET21a-PluAs-1, pPET21a-PluAs-2, and pPET21a-PluAs-3, and the C-terminus of the recombinant expression protein has a fusion-expressed 6×His tag.
[0051] Design PCR primers Pul-F64 and Pul-R3342 to amplify PluAs-1 as follows:
[0052] Pul-F64: CCC GAATTC TGTAACGACAACAGTTCTTCACCCTCC (forward primer, containing Eco RI restriction site), that is, SEQ ID NO 5.
[0053] Pul-R3342: CCC GCGGCCGC CCCCAAGAAGCCACGGACAAACA (reverse prime...
Embodiment 3
[0062] Example 3: Analysis of Enzymatic Characteristics of Recombinantly Expressed Pullulanase
[0063] (1) Determination of the optimum pH value: Prepare buffers with pH 3.0-10.0 containing 1% pullulan, among which pH 3.0-6.0 is 20mM citric acid-trisodium citrate buffer, and pH 7.0 is 20mM NaH 2 PO 4 -Na 2 HPO 4 Buffer, pH 8.0~10.5 is 20mM Tris-HCl buffer, pH 11.0 is 20mM glycine-NaOH buffer. Take 98 μL of the buffer solution containing the substrate and 2 μL of the purified enzyme solution and react in a water bath at 60°C for 10 min. Determination of pullulanase activity, wherein the highest enzyme activity is set to 100% (such as Figure 5 ).
[0064] (2) Determination of the optimum reaction temperature: Mix 2 μL of the purified enzyme solution with 98 μL of Tris-HCl containing 1% pullulan (pH 8.0), and mix them at 20, 30, 40, 50, 60 , reacted at 70 and 80 °C for 10 min, and then measured the activity of pullulanase by DNS method, among which the highest enzyme ac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com