Method for extracting genomic DNA (deoxyribonucleic acid) in milk appropriate for large-scale genotype identification
An extraction method, a large-scale technology, applied in the direction of recombinant DNA technology, DNA preparation, biochemical equipment and methods, etc., can solve the problems of large sampling volume, unsuitable for identification and molecular biological analysis, etc., to achieve convenient sampling and band The effect of uniformity and quality improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
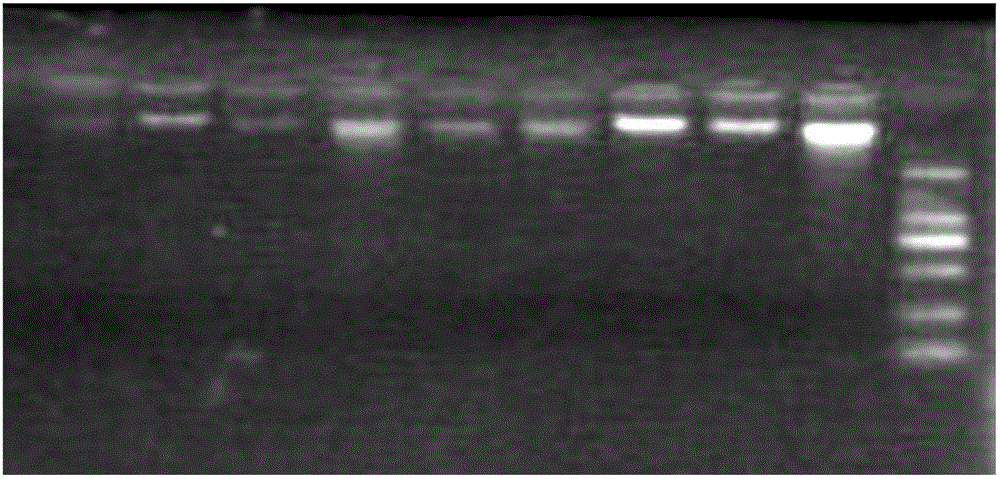
[0043] Embodiment 1: Extraction and quality detection of DNA in milk
[0044] 1. Separation and enrichment of milk somatic cells
[0045] The above cryopreserved milk sample was thawed at 4°C, and centrifuged at 2500r / min, 4°C for 30min. Use a small spoon to scrape off the milk fat on the upper layer of the centrifuge tube, and use a straw to remove the milk protein in the middle layer, leaving the bottom layer of sediment. Add 600 μL of PBS to the bottom of the centrifuge tube, beat the bottom pellet to suspend and transfer it to a 1.5 mL centrifuge tube, centrifuge at 3500 r / min for 10 min at room temperature, discard the upper liquid, and keep the bottom pellet. Then add 60 μL of emulsifier and 540 μL of PBS to the bottom sediment, shake with an oscillator until the sediment is completely suspended, treat it in a constant temperature water bath at 40°C for 10 minutes to remove the milk fat around the somatic cells, centrifuge at 3500 r / min for 10 minutes at room temperatur...
Embodiment 2
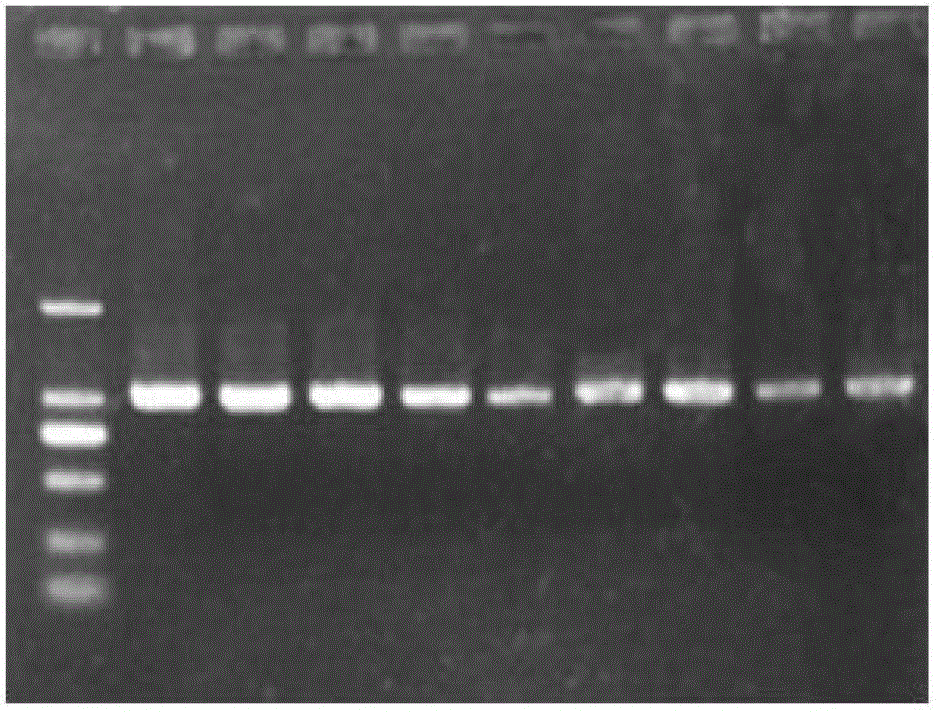
[0054] Example 2: PCR amplification and genotype identification of bovine B2M gene B2M1F and B2M1R primers
[0055] 1. Selection of specific genes
[0056] The bovine B2 microglobulin gene B2M (beta-2-microglobulin) was randomly selected, and its gene sequence number in GenBank is NC_007308.
[0057] 2. Design and synthesis of primers
[0058] Primers were designed according to the bovine B2M gene sequence and synthesized by Shanghai Sangon Bioengineering Company:
[0059] Upstream primer (B2M1F): 5'CAT CTG TCT TTC CCT GCC GC 3'
[0060] Downstream primer (B2M1R): 5' CTA CAG CCT TCC TCA TCT CCC CT 3'.
[0061] 3. PCR reaction
[0062] (1) Carry out PCR amplification using bovine DNA as a template, and the 10 μL reaction system contains the following solutions or reagents:
[0063]
[0064] (2) Mix the above solutions, and after repeated exploration of the PCR annealing temperature, 65°C is finally determined as the annealing temperature. The PCR reaction conditions are...
Embodiment 3
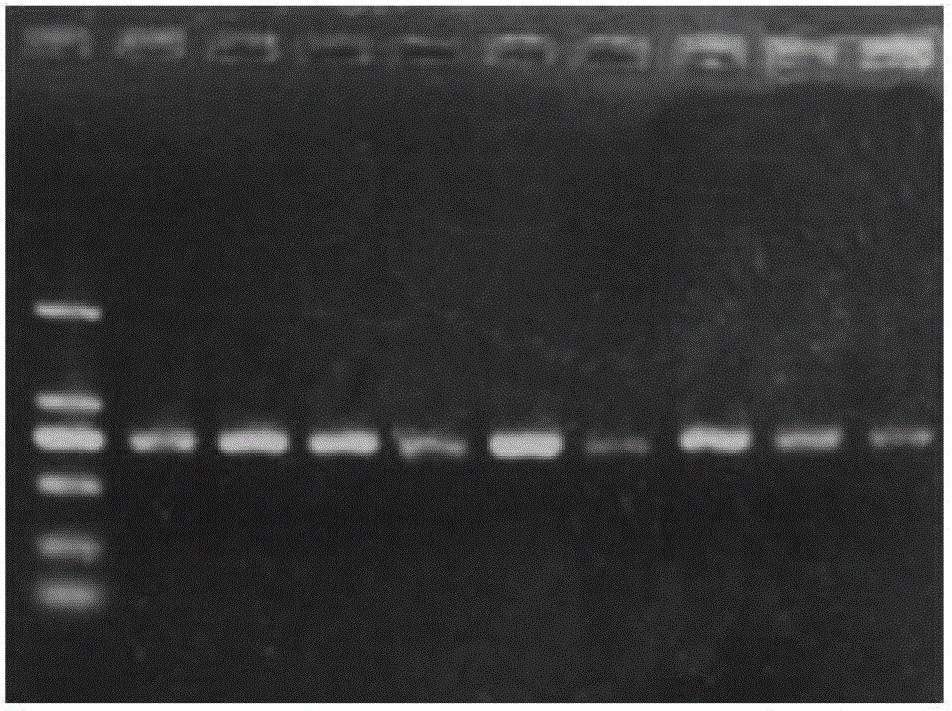
[0071] Example 3: PCR amplification and genotype identification of bovine B2M gene B2M2F and B2M2R primers
[0072] 1. Design and synthesis of primers
[0073] Primers were designed according to the bovine B2M gene sequence and synthesized by Shanghai Sangon Bioengineering Co., Ltd., wherein:
[0074] Upstream primer (B2M2F): 5'GGC TTT CCC AGC ATC ACT AAC 3'
[0075] Downstream primer (B2M2R): 5' TCA CAG CAC CAC CAA ACT TAT CT 3'.
[0076] 2. PCR reaction
[0077] (1) Carry out PCR amplification using bovine DNA as a template, and the 10 μL reaction system is the same as in Example 2.
[0078] (2) Mix the solutions of the PCR reaction system. After repeated exploration of the PCR annealing temperature, 60°C is finally determined as the annealing temperature. The PCR reaction conditions are as follows:
[0079]95°C, pre-denaturation for 5min; 94°C, denaturation for 30sec, 60°C, annealing for 30sec, 72°C extension for 30sec, 30 cycles; 72°C extension for 10min.
[0080] (3)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com