Method for preparing feed nucleotide by waste beer yeast
A technology of beer waste yeast and nucleotides, applied in the direction of microorganism-based methods, biochemical equipment and methods, animal feed, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
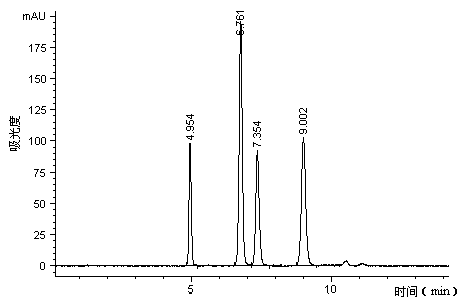
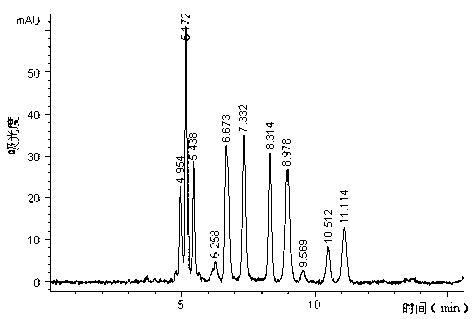
[0022] (1) Yeast wall breaking and RNA enzymatic hydrolysis: Add 8L of beer waste yeast sludge (equivalent to 890g of dry yeast) into a 10L reaction tank, dissolve 90g of solid sodium hydroxide in a small amount of water, and add it to the yeast under stirring Finally, add water to 9L, heat up to 40°C to break the wall of beer waste yeast and extract RNA for 2 hours, then add hydrochloric acid to the extract to make the pH of the extract 6.5, and take samples through the following UV The content of RNA in the extract was determined by spectrophotometry to be 4.14 mg / ml. According to the RNA content obtained by extraction, 2500-5000 activity units of 5'-phosphodiesterase were required per gram of RNA substrate, and determined and added 1460mL of 5'-phosphodiesterase enzyme solution, then adjust the pH value of the enzymolysis solution at 5.6-5.8, control the temperature at 65-70°C, and carry out the enzymolysis reaction for 3 hours; the 5'-nucleus is measured by the following p...
Embodiment 2
[0051] Take the fresh waste yeast sludge from the brewery, and the measured water content is 89%. Weigh out 3636g of yeast sludge (equivalent to 400g of dry yeast), put it in a 10L stainless steel stirring reaction tank, weigh 40g of solid NaOH and dissolve it in water, and add it into the tank while stirring , make the total volume reach 4000ml, heat up to 25°C, continue to stir and react for 3h, then adjust the pH to 6.5 with industrial hydrochloric acid, measure the content of RNA according to the method described in the above example 1, and the RNA substrate needs 5'- Calculation of phosphodiesterase 2500-5000 activity units, add 706ml of 5'-phosphodiesterase solution extracted from malt root, rapidly raise the temperature to 65°C, and enzymatically hydrolyze for 2 hours at the optimum pH 5.6 and optimum temperature 65°C , measure the 5'-nucleotide content according to the phosphorus determination method described in Example 1 above, and finally raise the temperature to 90°...
Embodiment 3
[0056] Take the fresh yeast mud from the brewery, and the measured water content is 89.6%. Measure 800L of fresh yeast mud into the tank through the standard barrel, which is equivalent to 83.2Kg of dry yeast. Add 27.6Kg of 30% NaOH under stirring, and the total volume is 830L. Heat up to 30°C, stir and react for 1.5h, then adjust the pH to 6.0 with 30% industrial hydrochloric acid, measure the content of RNA according to the method described in Example 1 above, and need 2500 g of 5'-phosphodiesterase per gram of RNA substrate Calculated at -5000 activity units, add 130L of 5'-phosphodiesterase solution extracted from malt root, rapidly raise the temperature to 70°C, maintain the optimum pH of 5.6-5.8 and the optimum temperature of 65-68°C for 3 hours, The 5'-nucleotide content was measured according to the phosphorus determination method described in Example 1 above, and finally the temperature was raised to 90° C. and maintained for 10 minutes to stop enzymatic hydrolysis.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com