SM gene for regulating and controlling cotton trichome, and application thereof
A gene and cotton technology, which is applied to the regulation of the SM gene of cotton wool and its application field, can solve the problems of rare application of cotton molecular markers and few applications.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1: SM gene precise positioning
[0020] 1) Creation of fine-mapping groups
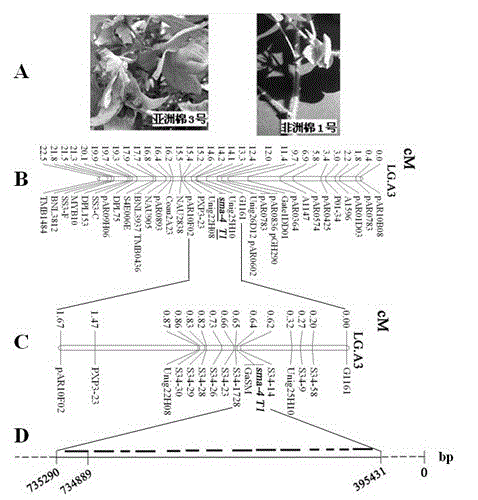
[0021] Use group A diploid African cotton No. 1 (A1, with fiber and hair, see figure 1 A) and group A diploid mutant Asian cotton No. 3 (A2, light seed, light rod, see figure 1 A) create F 2 Large population: 2366 strains [A2-113(s ma-4 )×A1-97 ( T 1 )] F 2 Mapping groups.
[0022] 2) Cotton genomic DNA extraction
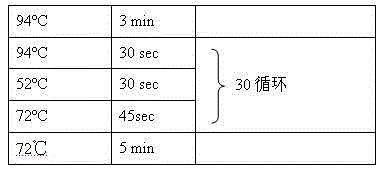
[0023] a) Take 0.2 g of fresh cotton leaves, put them into a mortar, add liquid nitrogen to grind to powder, quickly transfer to a 2.0 ml centrifuge tube, add 600 μl of preheated (65°C) CTAB extract (0.1 M Tris-HCl, pH8.0; 0.02M EDTA, pH8.0; 2% CTAB, 2% PVP40; 1 M sodium chloride; 0.2% β-mercaptoethanol), 65°C water bath for 60min, every 10min Shake once.
[0024] b) After the water bath, cool down to room temperature and add about 700ul of chloroform:isoamyl alcohol:absolute ethanol (volume ratio 76:4:20), mix well up and down, after static layering, centrifuge a...
Embodiment 2
[0037] Example 2: Obtaining the full-length sequence of the SM gene
[0038] 1) Obtain the full-length sequence of the gene using comparative genomics
[0039] According to the results of precise positioning and sequence comparison, it was determined that sma-4 / T1 Located in the third linkage group (LG.A3), and through the method of chromosome walking sma-4 / T1 limit Set within the interval of 340,000 bp. Determine the target gene by gene annotation, and design two sets of cross primers (5'-tcttctgaggtctgtctctg-3' / 5'-aaggcagcagctaagctatt-3' and 5'-attgcagcaacagcaagctc-3' and 5'- acttcaaacccctttctctc -3') to obtain two nucleic acid sequences covering the full length. After splicing and integrating the two sequences, the complete SM gene full-length genome sequence was obtained.
[0040] The full-length genome sequence of the group A diploid African cotton SM gene is shown in SEQ ID NO:4.
[0041] 2) Acquisition of full-length cDNA sequence
[0042] a) Extracti...
Embodiment 3
[0080] Embodiment 3: SM gene promoter sequence acquisition
[0081] Through the analysis of the reported cotton D genome Scaffold sequence, primers were designed to obtain the SM gene promoter sequence as shown in SEQ ID NO:3. Method is with embodiment 2.
[0082] Upstream primer: 5'-AATACCCTTTCATTGTCGATAAT-3'
[0083] Downstream primer: 5'-TTAACAAAGTGCTCGAATGTC-3'.
[0084] And it is shown through experiments that SM gene can control the production of fluff and affect fiber quality, and can be used to improve cotton fiber quality.
[0085]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com