Method for detecting enterovirus neutralizing antibody and special recombinant virus for method
A technology of enterovirus and recombinant virus, which is applied in the direction of virus/bacteriophage, chemical instruments and methods, botany equipment and methods, etc., can solve the problems of high cost, cumbersome operation, and danger, and achieve simple cost, easy standardization, The effect of easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
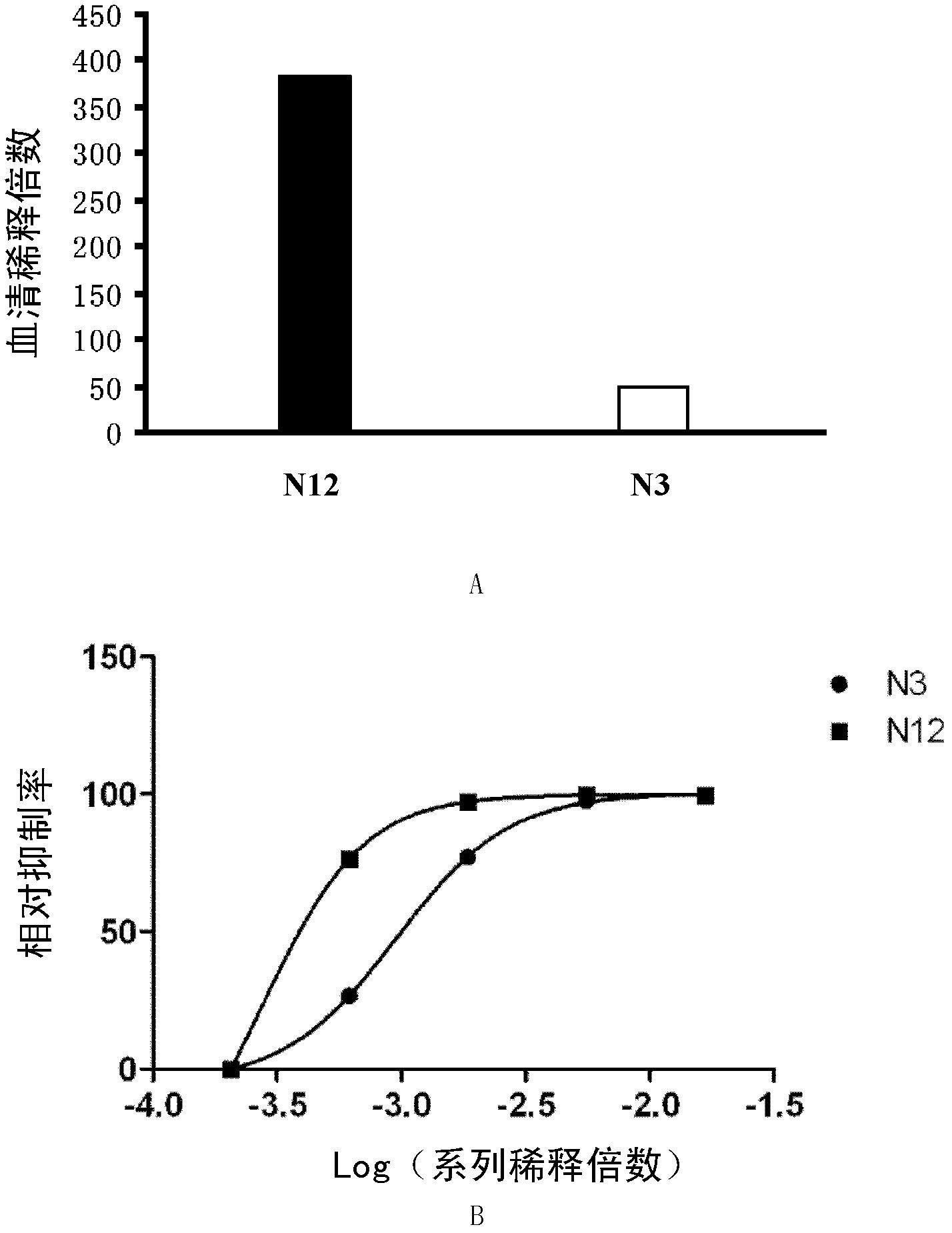
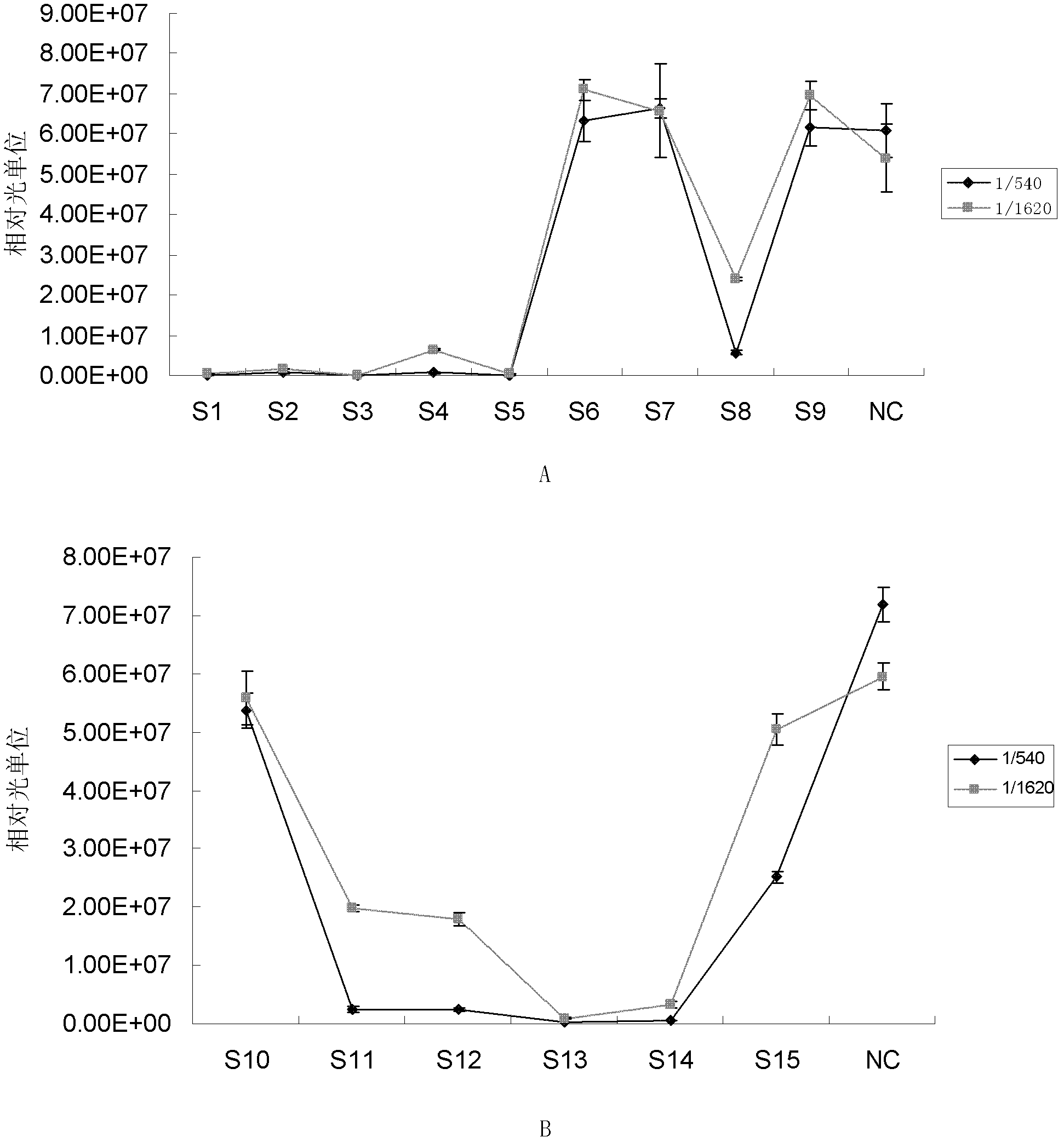
[0066] Embodiment 1, detect the neutralizing antibody of enterovirus EV71
[0067] 1. The effectiveness of qualitative detection of neutralizing antibodies against enterovirus EV71 and the titer of quantitative detection of neutralizing antibodies against enterovirus EV71
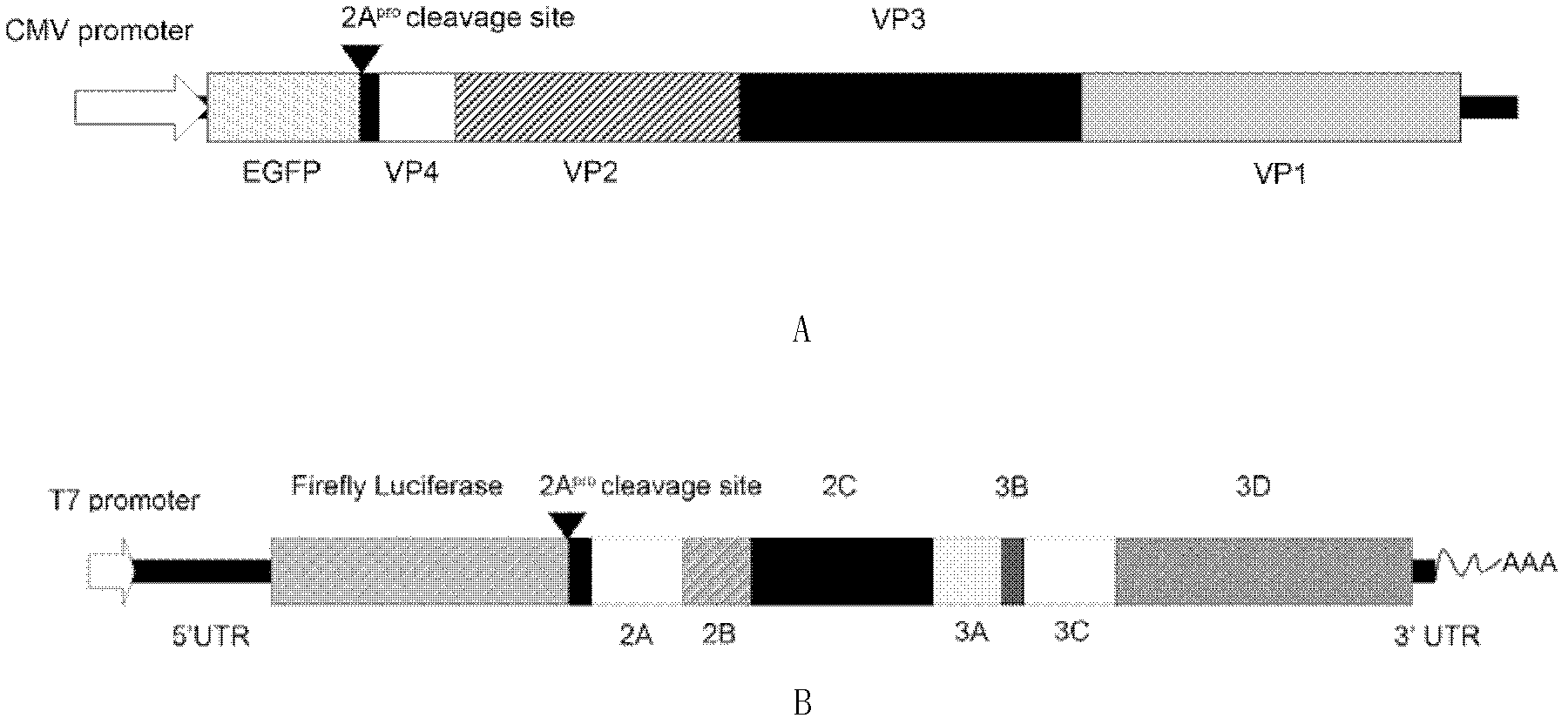
[0068] 1. Construction of EV71FY structural protein expression plasmid and EV71FY replicon plasmid
[0069] (1) Preparation of EV71 FY cDNA
[0070] Get the live virus EV71 / Fuyang.Anhui.P.R.C of EV71 FY (the public can obtain from Beijing Institute of Life Sciences, and the non-patent literature that has recorded this material is: Zhang, Y., Zhu, Z., Yang, W., Ren , J., Tan, X., Wang, Y., Mao, N., Xu, S., Zhu, S., Cui, A., Zhang, Y., Yan, D., Li, Q., Dong , X., Zhang, J., Zhao, Y., Wan, J., Feng, Z., Sun, J., Wang, S., Li, D. and Xu, W. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol. J. 7, 94 (2010)) culture flu...
Embodiment 2
[0120] Embodiment 2, detect the neutralizing antibody of Coxsackie virus A16
[0121] 1. Qualitative detection of the effectiveness of neutralizing antibodies against Coxsackievirus A16
[0122] 1. Construction of structural protein expression plasmids
[0123] (1) According to the DNA sequence of the structural protein gene of the G10 strain of Coxsackievirus A16 (NCBI sequence number: NC 001612), primers for whole gene synthesis were designed, and the gene was synthesized by PCR reaction. The primer sequences at both ends used in the whole gene synthesis are as follows:
[0124] CA16G10-F1-40: TGGACGAGCTGTACAAGGCCATTACTACCCTTGGGTCACA,
[0125] CA16G10-F2601-2624: AAAATAACAACACTATAAACCGGTAGCAGGTCAGTATGTG
[0126] (2. Construction of Cox A16 structural protein expression plasmid
[0127] The gene sequence of green fluorescent protein (EGFP) was spliced with the gene sequence of all structural proteins of Cox A16, and 2A pro The restriction site of protease is inserted i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com