Application of graphene in polymerase chain reaction as reinforcing agent
A technology of chain reaction and polymerase, applied in the field of graphene materials, to achieve the effect of simplifying steps, ensuring accuracy, and satisfying a large number of amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Mix 2g of natural graphite powder and 1g of sodium nitrate with 46mL of concentrated sulfuric acid in an ice bath, then add 6g of potassium permanganate, stir the mixture in an ice bath for 2 hours, then transfer to an oil bath at 35±5°C React for 30 minutes, the solution is brown and viscous, then slowly add 92mL of pure water to the mixture, raise the temperature to 95±5°C and continue the reaction for 3 hours, the mixture turns from brown to bright yellow, and finally add 6mL H with a mass fraction of 30% 2 O 2The solution is used to neutralize the unreacted potassium permanganate. After the above solution is cooled to room temperature, it is filtered with suction and the filter cake is repeatedly washed with pure water, dried in vacuum at 60° C. overnight to obtain graphite oxide. A certain mass of graphite oxide was weighed and dispersed into pure water, ultrasonicated for 5 hours to dissolve it completely, and then centrifuged at a low speed of 4000rpm...
Embodiment 2
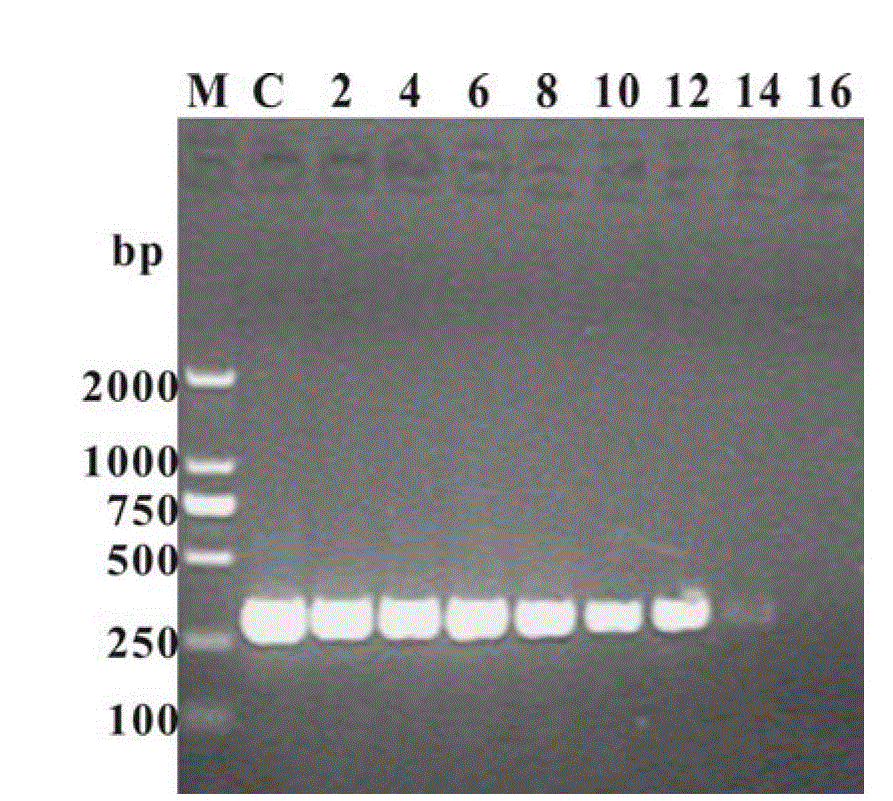
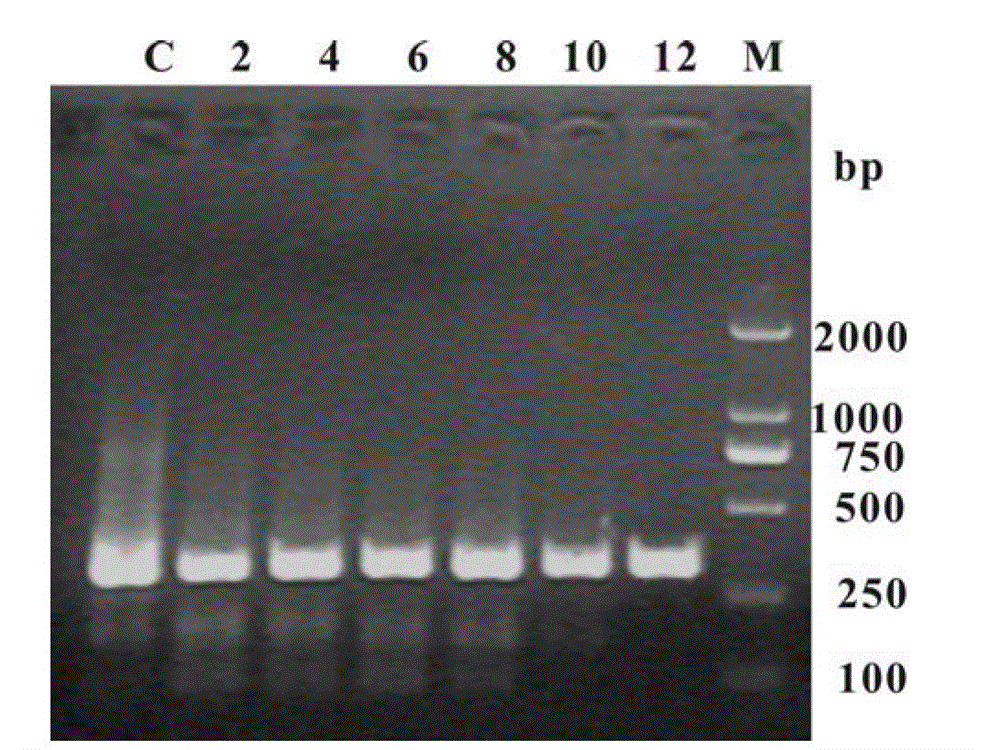
[0033] Example 2: The plasmid DNA template pET-32a was purchased from Novegan Company, and the length of the amplified target product was 300bp. The primer sequences used for amplification were: forward primer P15'-GCC ATG GAT CAC AAG CAT CAT GAT CAC-3', reverse primer P25'-GCC TCG AGG CAG AAA CAG TCG CAG T. The PCR system is 1×PCR buffer (20mM Tris-HCl, pH8.8, 10mM KCl, 16mM (NH 4 ) 2 SO 4 ,2mM MgSO 4 ,1~0.02mg mL -1 BSA, 0.1%TritonX-100), 0.2mM dNTPs, 0.4μM each of forward and reverse primers, 20ngμL -1 Plasmid DNA, 0.1U μL -1 PfuDNA polymerase, and finally the system volume was supplemented to 25 μl by graphene oxide or reduced graphene and sterilized double distilled water. In multiple rounds of PCR, the template DNA in each cycle was replaced by 0.4 μl of PCR product from the previous round. PCR conditions are: 1) pre-denaturation at 94°C for 5 min, 2) denaturation at 94°C for 30 s, annealing at 65°C for 30 s, extension at 72°C for 1 min, this step was repeated 30 ...
Embodiment 3
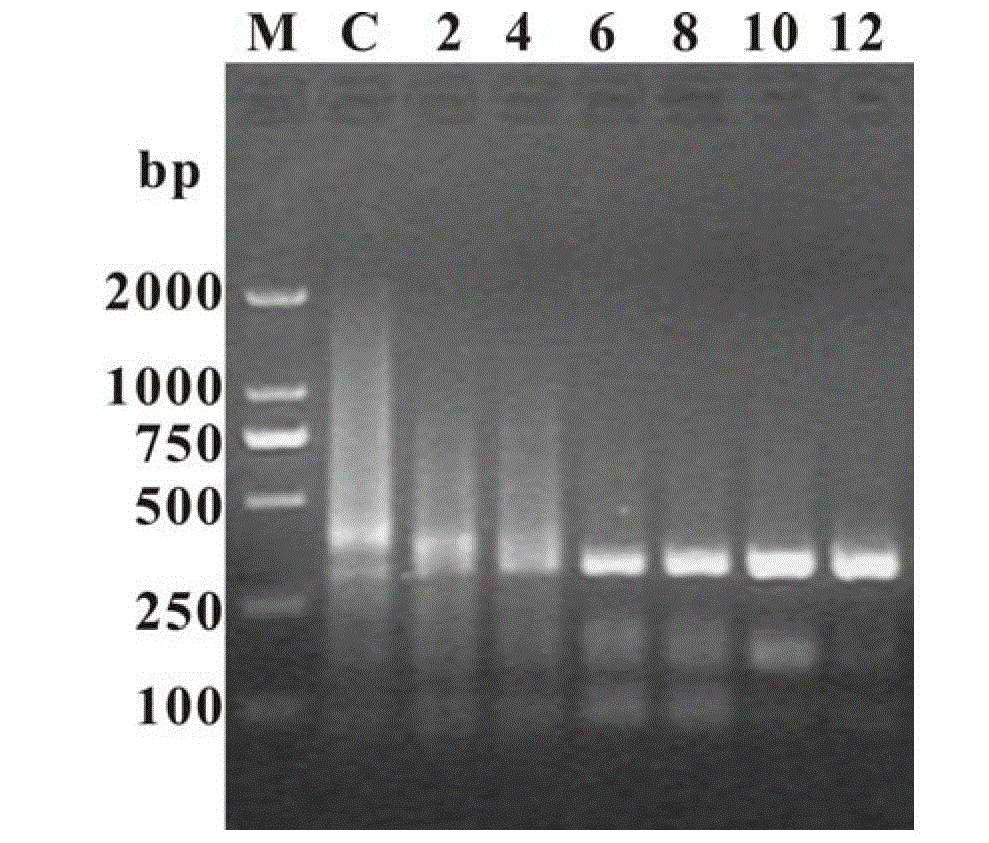
[0042] Example 3: The human genome template was collected from the blood of volunteers and used to amplify a 794bp DNA fragment located in the 12sRNA and 16sRNA regions of mitochondria. The amplification primers were forward primer 5'-AAC TAC GAT AGC CCT TATGAA-3', and reverse primer 5'-GGC CTACTATGG GTG TTAAA-3'. The PCR system is 1×PCRbuffer (20mM Tris-HCl, pH8.8, 10mM KCl, 16mM (NH 4 ) 2 SO 4 ,2mM MgSO 4 ,1~0.02mg mL -1 BSA, 0.1%TritonX-100), 0.2mM dNTPs, 0.4μM each of forward and reverse primers, 20ngμL -1 Genomic DNA, 0.1UμL -1 Pfu DNA polymerase, finally supplemented with graphene oxide or reduced graphene and sterilized double distilled water to make up the system volume to 25 μL. PCR conditions were 1) pre-denaturation at 94°C for 5 min, 2) denaturation at 94°C for 45 s, annealing at 52°C for 45 s, extension at 72°C for 90 s, this step was repeated 34 times, and 3) extension at 72°C for 10 min. Then use 1.5% EB stained agarose gel electrophoresis to detect the P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com