SiRNA (small interfering ribose nucleic acid) for inhibiting homosapiens chromosome2open reading frame 3(C2 or f3) (GCF) gene expression, carrier of SiRNA for inhibiting GCF gene expression and application
An expression vector and gene expression technology, applied in the field of medical genetic engineering, to achieve the effect of improving radiation sensitivity, benefiting radiotherapy and drug development, and high efficiency of RNA interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1 Screening of SiRNA sequences that inhibit GCF gene expression
[0065] Using network resources and software to conduct bioinformatics analysis on the GCF gene, search NCBI GeneBank to obtain the complete RNA sequence of the GCF gene, numbered NM_003203.4, with a total length of 4455bp, and its coding sequence (CDS, coding sequence) is located at 131-2476. Coding regions were chosen as target sequences for siRNA design. Perform preliminary design and screening on the corresponding siRNA design website (www.ambion.com), and then compare the following design principles to screen out the siRNA sequences. Principles of siRNA sequence design:
[0066] (1) 50-100 bases from the 5'-end of the promoter of the GCF gene.
[0067] (2) Find the 23 bases in the GCF gene sequence, preferably 5'-AA (N 19 )TT-3' (N is any base). If no more than four AAs are found (N 19 ) TT, use AA (N 21 ) make up. If no more than four AAs are found (N 21 ), then use NA (N 21 ) make u...
Embodiment 2
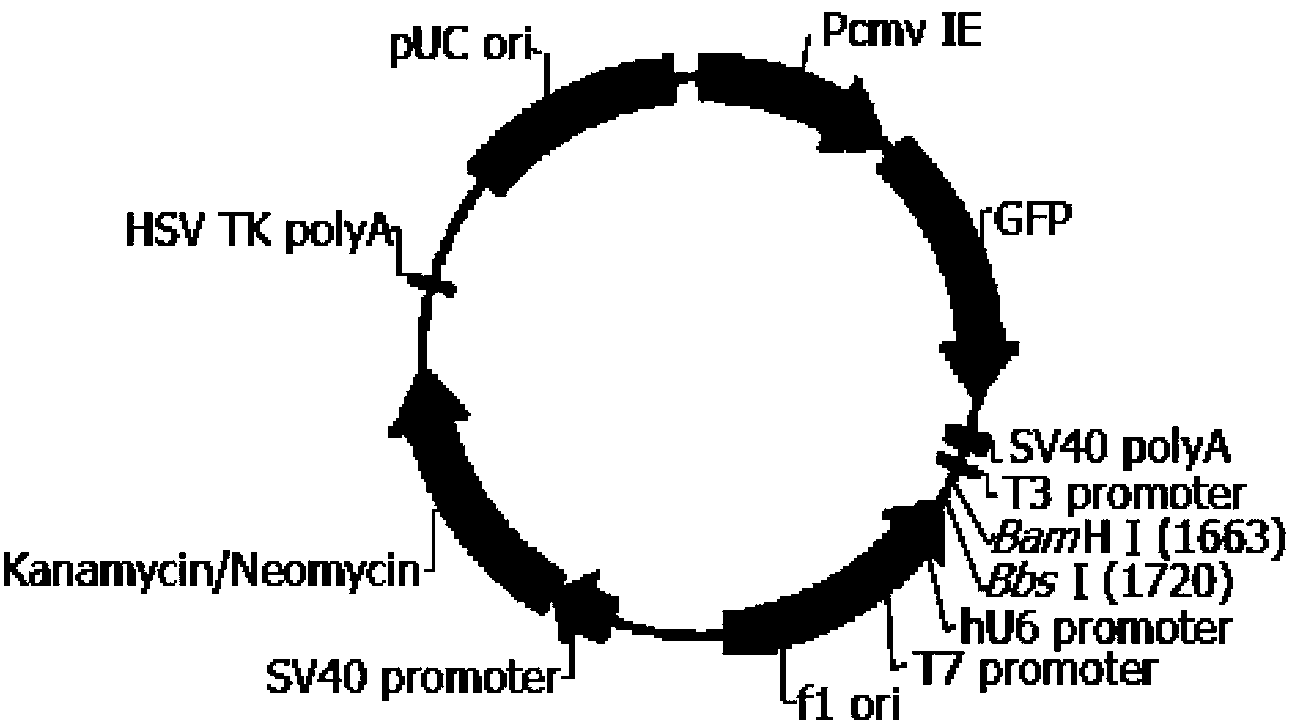
[0118] Embodiment 2 pGPU6 / GFP / Neo-shRNA vector construction
[0119] Including the following steps:
[0120] 1. Annealing of GCF-shRNA template
[0121] (1) GCF-shRNA796, GCF-shRNA1405, GCF-shRNA1696, GCF-shRNA1738 template sequences, positive control hGAPDH-shRNA template sequences and negative control NC-shRNA template sequences were artificially synthesized by Invitrogen, the sense strand or antisense The strand sequence is a hairpin structure.
[0122] (2) Dissolve the synthesized DNA oligo with TE (PH8.0) at a concentration of 100 μM.
[0123] (3) Anneal the sense strand and antisense strand of the hairpin structure to form a complementary double strand: the reaction system is 50 μL, where
[0124] Sense strand (100μM): 5μL; antisense strand (100μM): 5μL; annealing buffer (10×shDNAAnnealing Buffer): 5μL; ddH 2 O: 35 μL.
[0125] Perform annealing treatment on the PCR instrument according to the following procedures: 95°C, 5min; 85°C, 5min; 75°C, 5min; 70°C, 5min; St...
Embodiment 3
[0144] Example 3 Establishment and screening of pGPU6 / GFP / Neo-shRNA transfected HeLa cell line
[0145] pGPU6 / GFP / Neo-shRNA796 (named plasmid No. 1), pGPU6 / GFP / Neo-shRNA1405 (named plasmid No. 2), pGPU6 / GFP / Neo-shRNA1696 (named plasmid No. 3) constructed in Example 2 , pGPU6 / GFP / Neo-shRNA1738 (named plasmid No. 4), positive control pGPU6 / GFP / Neo-hGAPDH-shRNA (named plasmid No. 5), and negative control pGPU6 / GFP / Neo-NC-shRNA (named plasmid 6 No. plasmid) was transfected into human normal Hela cells to establish a monoclonal cell line, the specific steps are as follows:
[0146] 1. Determine the lethal dose of G418 for HeLa (normal human Hela cells were purchased from the Fourth Laboratory of the Second Academy of Military Medical Sciences)
[0147] The pGPU6 / GFP / Neo vector was purchased from Shanghai Gemma Company. It contains the anti-G418 gene and expresses green fluorescent protein. Therefore, these two indicators are used to screen successfully transfected monoclonal cells...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com