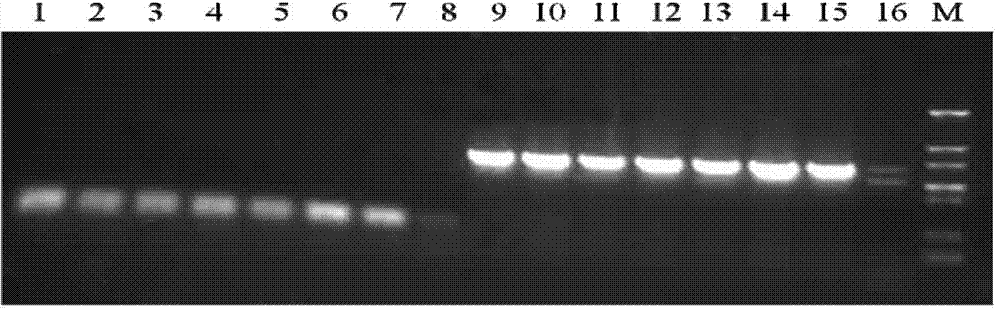
Method for efficiently expressing antibacterial peptide NZ2114 in recombinant pichia pastoris
A technology of NZ2114 and Pichia pastoris, which is applied in the field of genetic engineering, can solve problems such as no gene cloning and expression research reports, achieve broad application value and market prospects, and overcome the effect of low yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] The acquisition of embodiment 1 antimicrobial peptide NZ2114 gene fragment
[0062] 1.1 Optimization of gene expression sequence of antibacterial peptide NZ2114
[0063] According to the yeast codon table Pichia pastoris[gbpln]:137CDS's(81301codons):
[0064]
[0065]
[0066] According to the AA sequence of antimicrobial peptide NZ2114: GFGCNGPWNEDDLRCHNHCKSIKGYKGGYCAKGGFVCKCY (SEQ ID No.1), the gene encoding antimicrobial peptide NZ2114 was designed, and the DNA sequence (rNZ2114) after codon optimization was as follows:
[0067] GGT TTT GGT TGT AAC GGT CCA TGG AAC GAA GAT GATTTG AGA TGT CAT AAC CAT TGT AAG TCT ATT AAG GGT TACAAG GGT GGT TAC TGT GCT AAG GGT GGT TTT GTT TGT AAGTGT TAC (SEQ ID No. 2).
[0068] Sequence feature: 149bp; type: nucleic acid; chain type: double-stranded; topology: linear; GC%: 40.9%. Molecule Type: Double-stranded DNA.
[0069] 1.2 Design of gene expression cassette
[0070] In order to effectively terminate the translation express...
Embodiment 2
[0097] Example 2 Construction of Yeast Recombinant Expression Vector
[0098] 1.1 The NZ2114 gene fragment that embodiment 1 obtains reclaims transformation fragment after XhoI and XbaI endonuclease double digestion. At the same time, the pPICZαA vector (purchased from Invitrogen) was double digested with XhoI and XbaI.
[0099] The double enzyme digestion system is as follows:
[0100]
[0101] Digestion conditions: 37°C water bath for 4h.
[0102] The digested product was recovered with a DNA product recovery kit (purchased from Tiangen Biological Co., Ltd.), and stored at -20°C for future use. After the NZ2114 gene and the pPICZαA vector were digested with XbaI and XhoI, the NZ2114 gene was ligated with the linearized pPICZαA vector with T4 DNA ligase. The connection system is as follows:
[0103]
[0104] Connection conditions: 25°C, 1h.
[0105] 1.2 Transform the obtained recombinant vector into Escherichia coli DH5α: Take out the competent cells of Escherichia...
Embodiment 3
[0126] Example 3 Preparation of recombinant yeast strain X-33 containing NZ2114 gene
[0127] 1.1 Linearization of recombinant pPICZαA-NZ2114 vector
[0128] The recombinant pPICZαA-NZ2114 vector obtained in Example 2 was linearized with BglII endonuclease and used for yeast transformation.
[0129] The linearization system is as follows:
[0130]
[0131] Reaction conditions: 37°C, 4h.
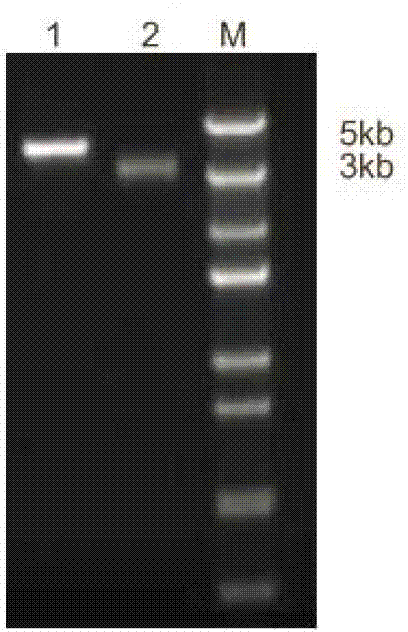
[0132] Electrophoresis results ( image 3 ) shows that the pPICZαA-NZ2114 recombinant vector is completely linearized.
[0133] 1.2 Preparation of competent Pichia pastoris X-33
[0134] Pick a single colony of Pichia pastoris X-33 (purchased from Invitrogen) and inoculate it into 10ml of YPD medium, shake it at 250rpm at 30°C overnight, and inoculate the overnight culture of Pichia pastoris X-33 into 50ml of YPD with 1% inoculum Medium, 250rpm, 30°C shaking culture to OD 600nm =1.3-1.5, 4°C 4000rpm, centrifuge for 5min, remove the supernatant, resuspend the bacteria in 50ml ice-cold...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com