Polymerase chain reaction (PCR) detection method of specificity of botrytis cinerea
A detection method and technology for Botrytis cinerea, which is applied in the field of microbial detection, can solve the problems of long PCR reaction time and limited to microgram level, etc., and achieve the effects of simple result determination, short detection time and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The present invention designs a kind of Botrytis cinerea specific PCR detection method, comprises the steps:
[0033] Step 1, designing primer pairs for specific amplification of Botrytis cinerea
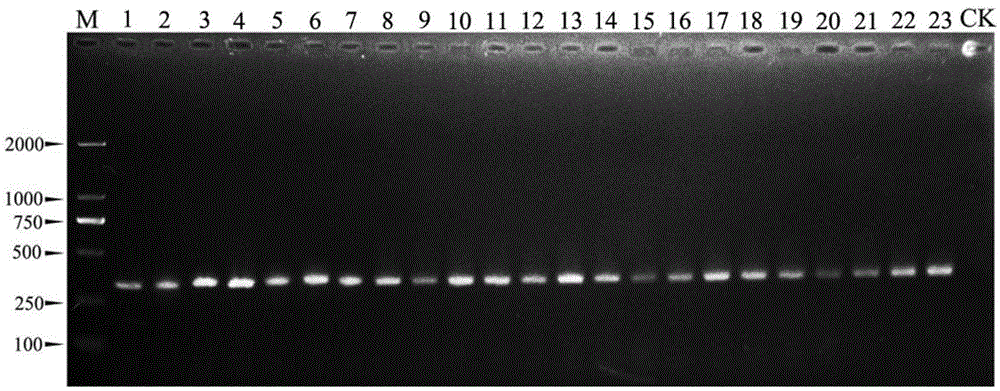
[0034] By searching the literature and referring to the RAPD (randomly amplified polymorphic DNA fragment) primers provided by Shanghai Sangon Bioengineering Co., Ltd., 23 random primers were selected (see Table 1, the sequences were synthesized by Beijing Aoke Dingsheng Biotechnology Co., Ltd.) , amplified using the genomic DNA of Botrytis cinerea and its relatives (see Table 3) as a template.
[0035] 50μL RAPD reaction system: sample genomic DNA 1μL (40ng); 10×PCR buffer 5μL; 25mM MgCl 2 2 μL; 2.5mM dNTP Mixture (TAKARA Biotechnology Co., Dalian) 1.6 μL; 5U / μL rTaq enzyme (TAKARA Biotechnology Co., Dalian) 0.5 μL; 20 μM RAPD random primer 3 μL; ddH 2 O36.9 μL. RAPD reaction program: pre-denaturation at 94°C for 4 min; denaturation at 94°C for 30 s, annealing at 36°C for...
Embodiment 2
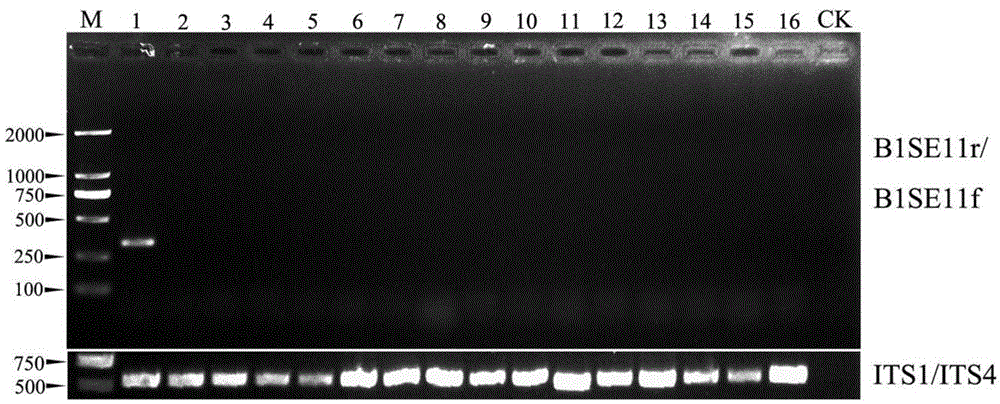
[0053] Specificity evaluation experiment of botrytis cinerea PCR detection method
[0054] Step 1, DNA template preparation
[0055] According to step 2 of Example 1, the genomic DNAs of 16 strains (see Table 3) of other fungi of the genus Botrytis and their related genera were respectively extracted.
[0056] The strains used in the present invention and their identification methods are the fungi that have been identified in the reference literature (Zhang Jing. Studies on Botrytis cinerea flora and Botrytis cinerea diversity in Hubei Province. Huazhong Agricultural University, doctoral dissertation, 2010). All experimental strains came from the research group of Phytopathogenic Fungus Li Guoqing of the State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University. At the same time, the present invention amplifies part of the eukaryotic ITS1-5.8S-ITS2 (eukaryotic ribosome transcribed spacer) region based on the universal primer pair ITS1 and ITS4 to ver...
Embodiment 3
[0066] Amplification of different concentrations of Botrytis cinerea DNA to evaluate the sensitivity of Botrytis cinerea PCR detection method
[0067] Step 1, DNA template preparation
[0068] According to step 2 of Example 1, extract Botrytis cinerea DNA, ddH 2 O was dissolved and diluted, and the concentration detected by the ultraviolet spectrophotometer was 40ng / μL, 20ng / μL, 4ng / μL, 0.8ng / μL, 0.4ng / μL, and 0.04ng / μL.
[0069] Step 2: Sensitivity evaluation test of Botrytis cinerea PCR detection method
[0070] Take 1 μL of each concentration sample and add it into the PCR reaction system, and perform PCR amplification detection on the DNA template to be tested according to the method in Step 3 of Example 1.
[0071] Step 3, result judgment
[0072] Take 10 μL of the PCR amplification product, detect it by 1.5% agarose gel electrophoresis, stain with EB, and observe under ultraviolet light. If there is a single amplification band at 327 bp, it means that the sample conta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com